Abstract
Background
The use of venom fractions from the Iranian cobra could be useful adjunct treatments of malaria with chloroquine. A metabolomic investigation with 1HNMR spectroscopy was conducted on an effective fraction tested earlier using Plasmodium berghei as an experimental murine model.
Purpose
We sought to ascertain both safety and anti-parasitic effects of experimental therapies.
Methods
After purification of the venom fractions, 25 mice were infected, then treated for 4 days with 0.2 ml of 5 mg/kg, 2.5 mg/kg and 1 mg/kg of the effective fraction, chloroquine, and a drug vehicle. An ED50 was obtained using Giemsa staining and real-time PCR analysis. The toxicity tests inspecting both liver and kidney tissues were performed.
Results
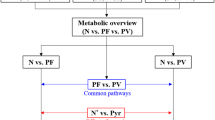
A clear inhibitory effect on parasitaemia was observed (with 75% inhibition with 5 mg/kg and 50% reduction when 2.5 mg/kg dosage used). ED50 obtained 2.5 mg/kg. The metabolomics were identified as differentiation of aminoacyl-t-RNA biosynthesis, valine, leucine, isoleucine biosynthesis and degradation pathways were observed.
Conclusion
Upon therapeutic effects of cobra venom fraction, further optimization of dose-dependent response of pharmacokinetics would be worthwhile for further exploration in adjunct experimental venom therapies.



Similar content being viewed by others
References
Oyelade J, Isewon I, Uwoghiren E, Aromolaran O, Oladipupo O (2018) In silico knockout screening of Plasmodium falciparum reactions and prediction of novel essential reactions by analysing the metabolic network. Biomed Res Int 2018:1–11. https://doi.org/10.1155/2018/8985718
Elmi T, Hajialiani F, Asadi MR, Orujzadeh F, Kalantari Hesari A, Rahimi Esboei B et al (2019) A study on the effect of Zingiber officinale hydroalcoholic extract on Plasmodium berghei in infected mice: an experimental study. JRUMS. 18(4):353–364
Garedaghi Y, Khaki A (2014) Sciences B, evaluation of the effectiveness of ethanolic extract of Solanum surattense against Plasmodium Berghei in comparison with chloroquine in Sourian mice using in vivo tests. Crescent J Med Biol Sci 1(3):76–79 (eISSN: 2148-9696)
Flegg JA, Guérin PJ, Nosten F, Ashley EA, Phyo AP, Dondorp AM et al (2013) Optimal sampling designs for estimation of Plasmodium falciparum clearance rates in patients treated with artemisinin derivatives. Malar J 12(1):411. https://doi.org/10.1186/1475-2875-12-411
Elmi T, Gholami S, Azadbakht M, Ziaei H (2014) Effect of chloroformic extract of Tanacetum parthenium in the treatment of Giardia lamblia infection in Balb/c mice. JMUMS 23(1):157–165
Vyas VK, Brahmbhatt K, Bhatt H, Parmar U (2013) Therapeutic potential of snake venom in cancer therapy: current perspectives. Asian Pac J Trop Biomed 3(2):156–162. https://doi.org/10.1016/S2221-1691(13)60042-8
Sang Chan Y, Chi Fai Cheung R, Xia L, Ho Wong J, Bun Ng T, Yee Chan W (2016) Snake venom toxins: toxicity and medicinal applications. Appl Microbiol Biotechnol 100(14):6165–6181. https://doi.org/10.1007/s00253-016-7610-9
Passero LF, Tomokane TY, Corbett CE, Laurenti MD, Toyama MH (2007) Comparative studies of the anti-leishmanial activity of three Crotalus durissus ssp. venoms. Parasitol Res 101(5):1365–1371. https://doi.org/10.1007/s00436-007-0653-1
Castillo J, Vargas L, Segura C, Gutiérrez J, Pérez JJT (2012) In vitro antiplasmodial activity of phospholipases A2 and a phospholipase homologue isolated from the venom of the snake Bothrops asper. Toxins (Basel). 4(12):1500–1516. https://doi.org/10.3390/toxins4121500
Maluf SEC, DalMas C, Oliveira EB, Melo P, Carmona AK, Gazarini ML et al (2016) Inhibition of malaria parasite Plasmodium falciparum development by crotamine, a cell penetrating peptide from the snake venom. Peptides 78:11–16. https://doi.org/10.1016/j.peptides.2016.01.013
Sandlers Y (2017) The future perspective: metabolomics in laboratory medicine for inborn errors of metabolism. Transl Res 189:65–75. https://doi.org/10.1016/j.trsl.2017.06.005
Klupczynska A, Pawlak M, Kokot ZJ, Matysiak J (2018) Application of metabolomic tools for studying low molecular-weight fraction of animal venoms and poisons. Toxins (Basel) 10(8):306. https://doi.org/10.3390/toxins10080306
Hajialiani F, Elmi T, Mohamadi M, Sadeghi S, Shahbazzadeh D, Ghaffarifar F, Dalimi A et al (2020) Analysis of the active fraction of Iranian Naja naja oxiana snake venom on the metabolite profiles of the malaria parasite by 1HNMR in vitro. Iran J Basic Med Sci 23:534–543. https://doi.org/10.22038/IJBMS.2020.39386.934
Nateghpour M, Edrissian G, Torabi A, Raesi A, Motevalli-Haghi A, Abed-Khojasteh H, Ghobakhlo N (2009) Monitoring of plasmodium vivax and plasmodium falciparum response to chloroquine in Bandar-Abbas district, Hormozgan province Iran. Tehran Univ Med J 67(3):178–183
Karbalaei Pazoki Z, Nateghpour M, Maghsood A, Souri E, Haghi AM, Farivar L et al (2014) Comparison between the effects of ethanolic extract of Artemisia annua and chloroquine on Plasmodium berghei in white mice. SJKUMS 19(2):9–20
World Health Organization. Regional Office for the Western Pacific (2016) Manila. https://iris.wpro.who.int/browse?type=author&;value=World%20Health%20Organization.%20Regional%20Office%20for%20the%20Western%20Pacific
Ebrahimzadeh A, Fouladi B, Fazaeli AJPI (2007) High rate of detection of mixed infections of Plasmodium vivax and Plasmodium falciparum in South-East of Iran, using nested PCR. Parasitol Int 56(1):61–64
White NJ (2004) Drug resistance. J Clin Invest 113(8):1084–1092
Uchida NS, Silva-Filho SE, Cardia GFE, Cremer E, Silva-Comar FMDS, Silva EL et al (2017) Hepatoprotective effect of citral on acetaminophen-induced liver toxicity in mice. Evid Based Complement Alternat Med 2017:1–9
Karbalaei Z, Nateghpour M, Maghsood A, Souri E (2014) Comparison between the effects of ethanolic extract of Artemisia annua and chloroquine on Plasmodium berghei in white mice. SJKU 19(2):9–20
Chutoam P, Klongthalay S, Somsak S (2015) Effect of crude leaf extract of Bauhinia strychnifolia in BALB/c mice infected with Plasmodium berghei. MCEJ 3(2):1–5
Khodaparast Z, Yousofi AR, Khoshvaghti A (2014) Investigation of curcumin effects on liver tissue in adult male rats treated with cyclophosphamide. JFUMS 4(3):344–352
Knight DJ, Peters W (1980) The antimalarial activity of Nbenzyloxy dihydrotriazines. The activity of clociguanil (BRL 50216) against rodent malaria, and studies on its mode of action. Ann Trop Med Parasitol 74(4):393–404 (PMID: 7006531)
Taylor SM, Juliano JJ, Trottman PA, Griffin JB, Landis SH, Kitsa P et al (2010) High-throughput pooling and real-time PCR-based strategy for malaria detection. JCM 48(2):512–519. https://doi.org/10.1128/JCM.01800-09
Zamani Z, Arjmand M, Vahabi F, IshaqHosseini SM, Fazeli SM, Sadeghi S et al (2014) A Metabolic study on colon cancer using 1H nuclear magnetic resonance spectroscopy. Biochem Res Int 2014:1–7. https://doi.org/10.1155/2014/348712
Zakeri S, Mamaghani S, Mehrizi A, Shahsavari Z, Raeisi A, Arshi S et al (2004) Molecular evidence of mixed P. vivax and P. falciparum infections in northern Islamic Republic of Iran. East Mediterr Health J 10(3):336–342 (PMID: 16212210)
Padley D, Moody A, Chiodini PL, Saldanha J (2003) Parasitology. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Ann Trop Med Parasitol 97(2):131–137. https://doi.org/10.1179/000349803125002977
Perandin F, Manca N, Piccolo G, Calderaro A, Galati L, Ricci L et al (2003) Identification of Plasmodium falciparum, P. vivax, P. ovale and P. malariae and detection of mixed infection in patients with imported malaria in Italy. New Microbiol 26(1):91–100
Richard FA, Sehgal RNM, Jones HI, Smith TB (2002) A comparative analysis of PCR-based detection methods for avian malaria. J Parasitol 88(4):819–822. https://doi.org/10.1645/0022-3395
Leopold SJ, Apinan S, Ghose A, Kingston HW, Plewes KA, Hossain A (2019) Amino acid derangements in adults with severe falciparum malaria. Sci Rep 9:6602. https://doi.org/10.1038/s41598-019-43044-6
Labaied M, Jayabalasingham B, Bano N, Cha SJ, Sandoval J, Guan G, Coppens I (2010) Plasmodium salvages cholesterol internalized by coA and synthesized de novo in the liver. Cell Microbiol. https://doi.org/10.1111/j.1462-5822.2010.01555.x
Oluba OM, Olusola AO, Eidangbe GO, Babatola LJ, Onyeneke EC (2012) Modulation of lipoprotein cholesterol levels in Plasmodium berghei malarial infection by crude aqueous extract of Ganoderma lucidum. Cholesterol 2012:1–6. https://doi.org/10.1155/2012/536396 (PMID: 22888413; PMCID: PMC3410371)
Enwonwu CO, Afolabi BM, Salako LO, Idigbe EO, Bashirelah N (2000) Increased plasma levels of histidine and histamine in falciparum malaria: relevance to severity of infection. J Neural Transm 107(11):1273–1287. https://doi.org/10.1007/s007020070017
Chukwuocha UM, Eke KN (2011) Malaria parasite status and cholesterol level of malaria patients in parts of the IMO River Basin of Nigeria Asian Pac. J Trop Med 4(12):993–996. https://doi.org/10.1016/S1995-7645(11)60232-0
Gal S, Fidler C, Turner S, Lo YD, Roberts DJ, Wainscoat JS (2001) Detection of Plasmodium falciparum DNA in plasma. Ann NY Acad 945(1):234–238. https://doi.org/10.1111/j.1749-6632.2001.tb03891.x
Ghosh S, Sengupta A, Sharma S, Sonawat HM (2012) Metabolic fingerprints of serum, brain, and liver are distinct for mice with cerebral and noncerebral malaria: a 1H NMR spectroscopy-based metabonomic study. J Proteome Res 11(10):4992–5004. https://doi.org/10.1021/pr300562m
Sengupta A, Ghosh S, Sharma S, Sonawat HM (2013) 1HNMR metabonomics indicates continued metabolic changes and sexual dimorphism post-parasite clearance in self-limiting murine malaria model. PLoS ONE 8(6):e66954. https://doi.org/10.1371/journal.pone.0066954
Adejuwon CA, Adejuwon AOJ (2005) Serum steroid levels in mice infected with Plasmodium berghei berghei. JMS 5(3):212–215
Istvan ES, Dharia NV, Bopp SE, Gluzman I, Winzeler EA, Goldberg DE (2011) Validation of isoleucine utilization targets in Plasmodium falciparum. PNAS 108(4):1627–1632. https://doi.org/10.1073/pnas.1011560108
Martin RE, Kirk KJB (2007) Transport of the essential nutrient isoleucine in human erythrocytes infected with the malaria parasite Plasmodium falciparum. Blood 109(5):2217–2224. https://doi.org/10.1182/blood-2005-11-026963
Bozdech Z, Ginsburg HJMJ (2005) Data mining of the transcriptome of Plasmodium falciparum: the pentose phosphate pathway and ancillary processes. Malar J 4(1):17. https://doi.org/10.1186/1475-2875-4-17
Shanmugasundram A, Gonzalez-Galarza FF, Wastling JM, Vasieva O, Jones RA (2013) Library of Apicomplexan metabolic pathways: a manually curated database for metabolic pathways of apicomplexan parasites. Nucleic Acids Res 41(12):D706–D713. https://doi.org/10.1093/nar/gks1139
Spry C, Saliba KJ (2009) The human malaria parasite Plasmodium falciparum is not dependent on host coenzyme A biosynthesis. J Biol Chem 84(37):24904–24913. https://doi.org/10.1074/jbc.M109.025312
Schalkwijk J, Allman EL, Jansen PA, de Vries LE, Verhoef JM, Jackowski S et al (2019) Antimalarial pantothenamide metabolites target acetyl–coenzyme A biosynthesis in Plasmodium falciparum. Sci Transl Med 11(510):9917. https://doi.org/10.1126/scitranslmed.aas9917
Lopansri BK, Anstey NM, Stoddard GJ, Mwaikambo ED, Boutlis CS, Tjitra E et al (2006) Elevated plasma phenylalanine in severe malaria and implications for pathophysiology of neurological complications. Infect Immun 74(6):3355–3359. https://doi.org/10.1128/IAI.02106-05
Müller S (2015) Role and regulation of glutathione metabolism in Plasmodium falciparum. Molecules 20(6):10511–10534. https://doi.org/10.3390/molecules200610511
Islahudin F, Pleass RJ, Avery SV, Ting K-NJJoac, (2012) Quinine interactions with tryptophan and tyrosine in malaria patients, and implications for quinine responses in the clinical setting. J Antimicrob Chemother 67(10):2501–2505. https://doi.org/10.1093/jac/dks253
Acknowledgements
We wish to acknowledge Iran National Science Foundation (INSF), Pasteur Institute of Iran and Iran University of Medical Sciences for jointly funding this Project.
Funding
This work was supported by Iranian University of Medical Sciences [Code: 9223651202].
Author information
Authors and Affiliations
Contributions
FH, ZZ, FT and TE performed experiments and analyzed the data. FT and FM searched the literature and designed the project. FT, ZZ and DS have participated in drafting the manuscript and supervised the research. FT and ZZ wrote the final manuscript. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethical Approval
This study was approved by the Ethics Committee of Iran University of Medical Sciences in accordance with guidelines. The designated approval code is: IR.IUMS.REC 1395.9223651202.
Conflict of Interests
The authors declare that there are no conflicts of interest.
Consent for Publication
The authors declare that they have no competing interests, and all agree to publish the paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11686_2021_456_MOESM1_ESM.docx
Supplementary file1 Supplement Fig.1 (A). The standard curve of cloned gene 18S rRNA sequence. (B) The red points show different concentration of standard gene, and the blue point show samples extracted from the mice for the ED50 test. Supplement Fig. 2 (A). The melt curve of the 18S rRNA gene synthesized and cloned into the PUC57 from samples extracted from the mice for the ED50 test. (B). The amplification of the 18S rRNA gene synthesized and cloned into the PUC57, and the samples extracted from the mice for the ED50 test. Supplement Fig.3. The histological sections of the liver for toxicity assay. Hematoxylin and Eosin staining with magnification of 100x, 400x and 420x. Number 1: A liver cell; Number 2: Central vein; Number 3: A macrophage or Kupffer cell. Negative control group: Hepatic lobules, the central vein, portal areas and hepatocytes were completely normal without hyperemia, edema or inflammation. (B) Positive control group: A slight degree of hyperemia was observed in this group. The invasion of the inflammatory cells, i.e., macrophages, was also visible in the liver parenchyma (shown with arrows). Explanation: The so-called Kupffer cells or macrophages in the liver tissue contain triangular or oblong nuclei from a histological perspective; nevertheless, the macrophages in the present histological section appeared to have recently entered the liver tissue from blood, and did not resemble Kupffer cells in terms of the appearance of their cytoplasm and nuclei. (C) ED50 effective fraction: Hyperemia was not observed in this group. The number of macrophages were also lower, and most of them appeared with a general structure of a Kupffer cell (shown with arrows). Supplement Fig.4. Histological sections of the kidney in the toxicity assay. Hematoxylin and Eosin staining with a magnification of upper row:40x, middle row:100x and lowest row:400x. Number 1: Renal corpuscle: Number 2: Proximal convoluted tubule; Number 3: Distal convoluted tubule. Negative control group: The histo-architecture of the cortex and center of the kidney was normal with no breakdown, hyperemia, edema and inflation. (B) Positive control group: In this group, the epithelial disruption of renal tubes was observed in some segments along with their release into the central lumen as well as sediments in proximal and distal convoluted tubules (shown with arrows). (C) ED50 effective fraction group: The entire histo-architecture of the kidney was normal in this group. Supplement Fig.5. Histological sections of the spleen in the toxicity assay. Hematoxylin and Eosin staining with a magnification of upper row 40x, middle row 100x and lower row 400x. Number 1: The white pulp of the spleen; Number 2: The red pulp of the spleen; Number 3: Lymphoid cells. Negative control group: The general histology of the spleen, including the white pulp comprising lymphatic cells and the red pulp comprising red blood cells, was normal with no hyperemia, edema, and inflation. (B) Positive control group: A significant influx of lymphatic cells, especially macrophages, was observable in this group. In some cases, the macrophages in this group were joined together forming giant cells (shown by arrows). (C) ED50 effective fraction: The entire histo-architecture of the spleen was normal in this group. Supplement Fig.6 (A). Score plot from chemometric analysis of spectra from ED50 of effective fraction and negative controls. (B). Score plot of spectra from ED50 of effective fraction and positive controls.Supplement Fig.7 (A). VIP scores from PLS-DA in ED50 of effective fractions and positive fractions showing levels of important components. (B). VIP scores from PLS-DA in ED50 of effective fractions and negative fractions showing levels of important components. (DOCX 1199 kb)
Rights and permissions
About this article
Cite this article
Hajialiani, F., Shahbazzadeh, D., Maleki, F. et al. The Metabolomic Profiles of Sera of Mice Infected with Plasmodium berghei and Treated by Effective Fraction of Naja naja oxiana Using 1H Nuclear Magnetic Resonance Spectroscopy. Acta Parasit. 66, 1517–1527 (2021). https://doi.org/10.1007/s11686-021-00456-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-021-00456-7