Abstract
Early diagnosis of hepatitis C virus (HCV) infection is essential to prevent disease from spreading and progression. Herein, a novel electrochemical biosensor was developed for ultrasensitive detection of HCV core antigen (HCVcAg) based on terminal deoxynucleotidyl transferase (TdT) amplification and DNA nanowires (DNW). After sandwich-type antibody-antigen recognition, the antibody-conjugated DNA was pulled to the electrode surface and further extended into a long DNA sequence by robust TdT reaction. Then, large numbers of methylene blue–loaded DNW (MB@DNW) as signal labels are linked to the extended DNA sequence. This results in an amplified electrochemical signal for HCVcAg determination, typically measured at around −0.25 V (Ag/AgCl). Under the optimum conditions, the proposed biosensor achieved a wide linear range for HCVcAg from 0.1 to 312.5 pg/mL with a low limit of detection of 32 fg/mL. The good practicality of the biosensor was demonstrated by recovery experiment (recoveries from 98 to 104% with RSD of 2.5–4.4%) and comparison with enzyme-linked immunosorbent assay (ELISA). Given the highlighted performance, the biosensor is expected to act as a reliable sensing tool for HCVcAg determination in clinics.
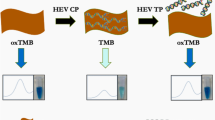
Graphical abstract

Schematic representation of the ultrasensitive electrochemical biosensor based on terminal deoxynucleotidyl transferase (TdT) amplification linked with methylene blue–loaded DNA nanowires (MB@DNW), which can be applied to the determination of hepatitis C virus core antigen (HCVcAg) in clinical samples. dTTPs, 2′-deoxythymidine 5′-triphosphate.







Similar content being viewed by others
References
Dore GJ, Martinello M, Alavi M, Grebely J (2020) Global elimination of hepatitis C virus by 2030: why not? Nat Med 26:157–160
Ge D, Fellay J, Thompson AJ (2009) Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401
Hanafiah KM, Groeger J, Flaxman AD, Wiersma ST (2013) Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342
Fabrizi F, Lunghi G, Aucella F, Mangano S, Barbisoni F, Bisegna S, Vigilante D, Limido A, Martin P (2005) Novel assay using total hepatitis C virus (HCV) core antigen quantification for diagnosis of HCV infection in dialysis patients. J Clin Microbiol 43:414–420
Seme K, Poljak M, Babic DZ, Mocilnik T, Vince A (2005) The role of core antigen detection in management of hepatitis C: a critical review. J Clin Virol 32:92–101
Bouvier-Alias M, Patel K, Dahari H, Beaucourt S, Larderie P, Blatt L, Hezode C, Picchio G, Dhumeaux D, Neumann AU, McHutchison JG, Pawlotsky JM (2002) Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology 36:211–218
Kaiser T, Damerow HC, Tenckhoff S, Finger A, Böttcher I, Hafer C, Schwarz A, Lüth JB, Schmidt Gürtler H, Colucci G, Manns MP, Wedemeyer H, Tillmann HL (2008) Kinetics of hepatitis C viral RNA and HCV-antigen during dialysis sessions: evidence for differential viral load reduction on dialysis. J Med Virol 80:1195–1201
Schüttler CG, Thomas C, Discher T, Friese G, Lohmeyer J, Schuster R, Schaefer S, Gerlich WH (2004) Variable ratio of hepatitis C virus RNA to viral core antigen in patient sera. J Clin Microbiol 42:1977–1981
Orito E, Mizokami M, Tanaka T, Lau JY, Suzuki K, Yamauchi M, Ohta Y, Hasegawa A, Tanaka S, Kohara M (1996) Quantification of serum hepatitis C virus core protein level in patients chronically infected with different hepatitis C virus genotypes. Gut 39:876–880
Valipour A, Roushani M (2017) TiO2 nanoparticles doped with Celestine Blue as a label in a sandwich immunoassay for the hepatitis C virus core antigen using a screen printed electrode. Microchim Acta 184:2015–2022
Valipour A, Roushani M (2017) A glassy carbon immunoelectrode modified with vanadium oxide nanobelts for ultrasensitive voltammetric determination of the core antigen of hepatitis C virus. Microchim Acta 184:4477–4483
Valipour A, Roushani M (2017) Using silver nanoparticle and thiol graphene quantum dots nanocomposite as a substratum to load antibody for detection of hepatitis C virus core antigen: electrochemical oxidation of riboflavin was used as redox probe. Biosens Bioelectron 89:946–951
Ghanbari K, Roushani M (2018) A nanohybrid probe based on double recognition of an aptamer MIP grafted onto a MWCNTs-Chit nanocomposite for sensing hepatitis C virus core antigen. Sens Actuators B-Chem 258:1066–1071
Li X, Yin C, Wu Y, Zhang Z, Jiang D, Xiao D, Fang X, Zhou C (2020) Plasmonic nanoplatform for point-of-care testing trace HCV core protein. Biosens Bioelectron 147:111488
Li X, Iocozzia J, Chen Y, Zhao S, Cui X, Wang W, Yu H, Lin S, Lin Z (2018) From precision synthesis of block copolymers to properties and applications of nanoparticles. Angew Chem Int Ed Eng 57:2046–2070
Xiao M, Lai W, Man T, Chang B, Li L, Chandrasekaran AR, Pei H (2019) Rationally engineered nucleic acid architectures for biosensing applications. Chem Rev 119:11631–11717
Wang W, Yu S, Huang S, Bi S, Han H, Zhang JR, Lu Y, Zhu JJ (2019) Bioapplications of DNA nanotechnology at the solid-liquid interface. Chem Soc Rev 48:4892–4920
Li F, Zhou Y, Yin H, Ai S (2020) Recent advances on signal amplification strategies in photoelectrochemical sensing of microRNAs. Biosens Bioelectron 166:112476
Zeng Z, Zhou R, Sun R, Zhang X, Cheng Z, Chen C, Zhu Q (2020) Nonlinear hybridization chain reaction-based functional DNA nanostructure assembly for biosensing, bioimaging applications. Biosens Bioelectron 173:112814
Wang H, Wang H, Willner I, Wang F (2020) High-performance biosensing based on autonomous enzyme-free DNA circuits. Top Curr Chem 378:20
Ge Z, Gu H, Li Q, Fan C (2018) Concept and development of framework nucleic acids. J Am Chem Soc 140:17808–17819
Huang X, Niu W, Wu J, Wang Y, Li C, Qiu J, Xue J (2019) A triple-amplification differential pulse voltammetry for sensitive detection of DNA based on exonuclease III, strand displacement reaction and terminal deoxynucleotidyl transferase. Biosens Bioelectron 143:111609
Wang L, Pan Y, Liu Y, Sun Z, Huang Y, Li J, Yang J, Xiang Y, Li G (2020) Fabrication of an aptamer-coated liposome complex for the detection and profiling of exosomes based on terminal deoxynucleotidyl transferase-mediated signal amplification. ACS Appl Mater Interfaces 12:322–329
Chen X, Cao G, Wang X, Ji Z, Xu F, Huo D, Luo X, Hou C (2020) Terminal deoxynucleotidyl transferase induced activators to unlock the trans-cleavage of CRISPR/Cpf1 (TdT-IU-CRISPR/Cpf1): an ultrasensitive biosensor for Dam MTase activity detection. Biosens Bioelectron 163:112271
Zheng J, Shi H, Wang M, Duan C, Huang Y, Li C, Xiang Y, Li G (2020) Homogenous electrochemical method for ultrasensitive detection of tumor cells designed by introduction of poly(A) tails onto cell membranes. Anal Chem 92:2194–2200
Lei S, Liu Z, Xu L, Zou L, Li G, Ye B (2020) A “signal-on” electrochemical biosensor based on DNAzyme-driven bipedal DNA walkers and TdT-mediated cascade signal amplification strategy. Anal Chim Acta 1100:40–46
Li X, Du Z, Lin S, Tian J, Tian H, Xu W (2020) ExoIII and TdT dependent isothermal amplification (ETDA) colorimetric biosensor for ultra-sensitive detection of Hg2+. Food Chem 316:126303
Liu Q, Ge Z, Mao X, Zhou G, Zuo X, Shen J, Shi J, Li J, Wang L, Chen X, Fan C (2018) Valency-controlled framework nucleic acid signal amplifiers. Angew Chem Int Ed Eng 57:7131–7135
Liu P, Qian X, Li X, Fan L, Li X, Cui D, Yan Y (2020) Enzyme-free electrochemical biosensor based on localized DNA Cascade displacement reaction and versatile DNA nanosheets for ultrasensitive detection of exosomal MicroRNA. ACS Appl Mater Interfaces 12:45648–45656
Zhao Y, Hu S, Wang H, Yu K, Guan Y, Liu X, Li N, Liu F (2017) DNA dendrimer-streptavidin nanocomplex: an efficient signal amplifier for construction of biosensing platforms. Anal Chem 89:6907–6914
Rahbani JF, Hariri AA, Cosa G, Sleiman HF (2015) Dynamic DNA nanotubes: reversible switching between single and double-stranded forms, and effect of base deletions. ACS Nano 9:11898–11908
Hariri AA, Hamblin GD, Gidi Y, Sleiman HF, Cosa G (2015) Stepwise growth of surface-grafted DNA nanotubes visualized at the single-molecule level. Nat Chem 7:295–300
Jiang Q, Song C, Nangreave J, Liu X, Lin L, Qiu D, Wang ZG, Zou G, Liang X, Yan H, Ding B (2012) DNA origami as a carrier for circumvention of drug resistance. J Am Chem Soc 134:13396–13403
Xue C, Zhang S, Yu X, Hu S, Lu Y, Wu ZS (2020) Periodically ordered, nuclease-resistant DNA nanowires decorated with cell-specific aptamers as selective theranostic agents. Angew Chem Int Ed Eng 59:17540–17547
Zong C, Wu J, Liu M, Yang L, Yan F, Ju H (2014) Chemiluminescence imaging for a protein assay via proximity-dependent DNAzyme formation. Anal Chem 86:9939–9944
Wang B, Shi S, Yang X, Wang Y, Qi H, Gao Q, Zhang C (2020) Separation-free electrogenerated chemiluminescence immunoassay incorporating target assistant proximity hybridization and dynamically competitive hybridization of a DNA signal probe. Anal Chem 92:884–891
Zhou Z, Xiang Y, Tong A, Lu Y (2014) Simple and efficient method to purify DNA-protein conjugates and its sensing applications. Anal Chem 86:3869–3875
Wang X, Gao H, Qi H, Gao Q, Zhang C (2018) Proximity hybridization-regulated immunoassay for cell surface protein and protein-overexpressing cancer cells via electrochemiluminescence. Anal Chem 90:3013–3018
Funding
This work was supported by the Natural Science Foundation of Chongqing (cstc2020jcyj-msxmX0764), the Combined Medical Funding of Chongqing Health Commission & Science and Technology Bureau (2020FYYX060), and the Xinglin Scholar Research Premotion Project of Chengdu University of TCM (YYZX2019067).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing of interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 179 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Li, L., Dong, Z. et al. Ultrasensitive electrochemical detection of hepatitis C virus core antigen using terminal deoxynucleotidyl transferase amplification coupled with DNA nanowires. Microchim Acta 188, 285 (2021). https://doi.org/10.1007/s00604-021-04939-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-04939-2