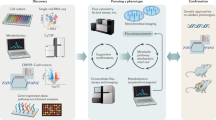
Abstract
T cells are integral players in the adaptive immune system that readily adapt their metabolism to meet their energetic and biosynthetic needs. A major hurdle to understand physiologic T-cell metabolism has been the differences between in vitro cell culture conditions and the complex in vivo milieu. To address this, we have developed a protocol that merges traditional immunology infection models with whole-body metabolite infusion and mass-spectrometry-based metabolomic profiling to assess T-cell metabolism in vivo. In this protocol, pathogen-infected mice are infused via the tail vein with an isotopically labeled metabolite (2–6 h), followed by rapid magnetic bead isolation to purify T-cell populations (<1 h) and then stable isotope labeling analysis conducted by mass spectrometry (~1–2 d). This procedure enables researchers to evaluate metabolic substrate utilization into central carbon metabolic pathways (i.e., glycolysis and the tricarboxylic acid cycle) by specific T-cell subpopulations in the context of physiological immune responses in vivo.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Cham, C. M., Driessens, G., O’Keefe, J. P. & Gajewski, T. F. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur. J. Immunol. 38, 2438–2450 (2008).
Chang, C. H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Ma, E. H. et al. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8+ T cells. Immunity 51, 856–870.e5 (2019).
Johnson, M. O. et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell (2018). https://doi.org/10.1016/j.cell.2018.10.001
Blagih, J. et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54 (2015).
Leone, R. D. et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 366, 1013–1021 (2019). https://doi.org/10.1126/science.aav2588
Balmer, M. L. et al. Memory CD8+ T cells require increased concentrations of acetate induced by stress for optimal function. Immunity 44, 1312–1324 (2016).
Qiu, J. et al. Acetate promotes T cell effector function during glucose restriction. Cell Rep. 27, 2063–2074.e5 (2019).
Ma, E. H. et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. 25, 345–357 (2017).
Davidson, S. M. et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 (2016).
Cantor, J. R. The rise of physiologic media. Trends Cell Biol. 29, 854–861 (2019).
Ackermann, T. & Tardito, S. Cell culture medium formulation and its implications in cancer metabolism. Trends Cancer 5, 329–332 (2019).
Voorde, J. V. et al. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci. Adv. 5, eaau7314 (2019).
Cantor, J. R. et al. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase HHS Public Access. Cell 169, 258–272 (2017).
Kaymak, I., Williams, K. S., Cantor, J. R. & Jones, R. G. Immunometabolic interplay in the tumor microenvironment. Cancer Cell 39, 28–37 (2020).
Hensley, C. T. et al. Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016).
Sellers, K. et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Invest. 125, 687–698 (2015).
Faubert, B. et al. Lactate metabolism in human lung tumors. Cell 171, 358–371.e9 (2017).
Yuan, M. et al. Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC–MS/MS. Nat. Protoc. 14, 313–330 (2019). https://doi.org/10.1038/s41596-018-0102-x
Roy, D. G., Kaymak, I., Williams, K. S., Ma, E. H. & Jones, R. G. Immunometabolism in the tumor microenvironment. Annu. Rev. Cancer Biol. 5, annurev-cancerbio-030518-055817 (2021).
Geltink, R. I. K., Kyle, R. L. & Pearce, E. L. Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 36, 461–488 (2018). https://doi.org/10.1146/annurev-immunol-042617-053019
Wang, R. & Green, D. R. Metabolic checkpoints in activated T cells. Nat. Immunol. 13, 907–915 (2012).
Pearce, E. L., Poffenberger, M. C., Chang, C. H. & Jones, R. G. Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454 (2013).
O’Sullivan, D. & Pearce, E. L. Targeting T cell metabolism for therapy. Trends Immunol. 36, 71–80 (2015).
Llufrio, E. M., Wang, L., Naser, F. J. & Patti, G. J. Sorting cells alters their redox state and cellular metabolome. Redox Biol. 16, 381–387 (2018).
Binek, A. et al. Flow cytometry has a significant impact on the cellular metabolome. J. Proteome Res. 18, 169–181 (2019).
Chen, W. W., Freinkman, E. & Sabatini, D. M. Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites. Nat. Protoc. 12, 2215–2231 (2017).
Szabo, P. A. et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. https://doi.org/10.1038/s41467-019-12464-3 (2019).
Miragaia, R. J. et al. Single-cell transcriptomics of regulatory t cells reveals trajectories of tissue adaptation. Immunity https://doi.org/10.1016/j.immuni.2019.01.001 (2019).
Lau, A. N. et al. Dissecting cell-type-specific metabolism in pancreatic ductal adenocarcinoma. eLife https://doi.org/10.7554/eLife.56782 (2020).
Fernández-García, J., Altea-Manzano, P., Pranzini, E. & Fendt, S. M. Stable isotopes for tracing mammalian-cell metabolism in vivo. Trends Biochem. Sci. https://doi.org/10.1016/j.tibs.2019.12.002 (2020).
Jang, C., Chen, L. & Rabinowitz, J. D. Metabolomics and isotope tracing. Cell https://doi.org/10.1016/j.cell.2018.03.055 (2018).
Andrikopoulos, S., Blair, A. R., Deluca, N., Fam, B. C. & Proietto, J. Evaluating the glucose tolerance test in mice. Am. J. Physiol. Endocrinol. Metab. https://doi.org/10.1152/ajpendo.90617.2008 (2008).
Tompkins, S. C. et al. Disrupting mitochondrial pyruvate uptake directs glutamine into the TCA cycle away from glutathione synthesis and impairs hepatocellular tumorigenesis. Cell Rep. https://doi.org/10.1016/j.celrep.2019.07.098 (2019).
Marin-Valencia, I. et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 15, 827–837 (2012).
Overmyer, K. A., Thonusin, C., Qi, N. R., Burant, C. F. & Evans, C. R. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: studies in a C57BL/6J mouse model. PLoS One 10, e0117232 (2015).
Guo, N. L., Zhang, J. X., Wu, J. P. & Xu, Y. H. Isoflurane promotes glucose metabolism through up-regulation of miR-21 and suppresses mitochondrial oxidative phosphorylation in ovarian scancer cells. Biosci. Rep. 37, BSR20170818 (2017).
Rao, L. K., Flaker, A. M., Friedel, C. C. & Kharasch, E. D. Role of cytochrome P4502B6 polymorphisms in ketamine metabolism and clearance. Anesthesiology 125, 1103–1112 (2016).
Kharasch, E. D. & Thummel, K. E. Identification of cytochrome P450 2E1 as the predominant enzyme catalyzing human liver microsomal defluorination of sevoflurane, isoflurane, and methoxyflurane. Anesthesiology 79, 795–807 (1993).
Quaegebeur, A. et al. Deletion or inhibition of the oxygen sensor PHD1 protects against ischemic stroke via reprogramming of neuronal metabolism. Cell Metab. https://doi.org/10.1016/j.cmet.2015.12.007 (2016).
Elia, I. et al. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. https://doi.org/10.1038/ncomms15267 (2017).
Jang, C. et al. The small intestine converts dietary fructose into glucose and organic acids. Cell Metab. https://doi.org/10.1016/j.cmet.2017.12.016 (2018).
Sun, R. C. et al. Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing. Nat. Commun. 8, 1646 (2017).
Goldberg, E. L. et al. Ketogenic diet activates protective γδ T cell responses against influenza virus infection. Sci. Immunol. https://doi.org/10.1126/sciimmunol.aav2026 (2019).
Dyar, K. A. et al. Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell 174, 1571–1585.e11 (2018).
Eckel-Mahan, K. L. et al. Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 (2013).
Kinouchi, K. et al. Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Rep. 25, 3299–3314.e6 (2018).
Shen, H. et al. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc. Natl Acad. Sci. USA 92, 3987–3991 (1995).
Shen, H. et al. Compartmentalization of bacterial antigens: differential effects on priming of CD8 T cells and protective immunity. Cell 92, 535–545 (1998).
Pope, C. et al. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J. Immunol. https://doi.org/10.4049/jimmunol.166.5.3402 (2001).
Agrawal, S. et al. El-MAVEN: a fast, robust, and user-friendly mass spectrometry data processing engine for metabolomics. Methods Mol. Biol. 1978, 301–321 (2019).
Badovinac, V. P., Haring, J. S. & Harty, J. T. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity 26, 827–841 (2007).
Best, J. A. et al. Transcriptional insights into the CD8 + T cell response to infection and memory T cell formation. Nat. Immunol. 14, 404–412 (2013).
Wang, Y. et al. Uncoupling hepatic oxidative phosphorylation reduces tumor growth in two murine models of colon cancer. Cell Rep. 24, 47–55 (2018).
Hui, S. et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017).
Tardito, S. et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat. Cell Biol. 17, 1556–1568 (2015).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Heinrich, P. et al. Correcting for natural isotope abundance and tracer impurity in MS-, MS/MS- and high-resolution-multiple-tracer-data from stable isotope labeling experiments with IsoCorrectoR. Sci. Rep. 8, 17910 (2018).
Trefely, S., Ashwell, P. & Snyder, N. W. FluxFix: automatic isotopologue normalization for metabolic tracer analysis. BMC Bioinformatics 17, 485 (2016).
Izreig, S. et al. Repression of LKB1 by miR-17∼92 sensitizes MYC-dependent lymphoma to biguanide treatment. Preprint at bioRxiv https://doi.org/10.1101/2019.12.20.883025 (2019).
Janzer, A. et al. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1409844111 (2014).
Spalding, J. L., Naser, F. J., Mahieu, N. G., Johnson, S. L. & Patti, G. J. Trace phosphate improves ZIC-pHILIC peak shape, sensitivity, and coverage for untargeted metabolomics. J. Proteome Res. 17, 3537–3546 (2018).
Mahieu, N. G. & Patti, G. J. Systems-level annotation of a metabolomics data set reduces 25 000 features to fewer than 1000 unique metabolites. Anal. Chem. 89, 10397–10406 (2017).
Yang, Q. et al. NOREVA: enhanced normalization and evaluation of time-course and multi-class metabolomic data. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa258 (2020).
Acknowledgements
We thank E. Levine, R. Sheridan and staff from the Van Andel Institute (VAI) Metabolomics and Bioenergetics and Flow Cytometry core facilities for technical assistance. We thank J. Wedberg and M. Minard for administrative assistance. Funding: The VAI Metabolomics and Bioenergetics Core Facility is supported by the Department of Metabolism and Nutritional Programming at VAI. This research has been supported by grants from the CIHR (MOP-142259 to RGJ) and funding from VAI and Agios Pharmaceuticals.
Author information
Authors and Affiliations
Contributions
Conceptualization: R.D.S., E.H.M. and R.G.J.; validation: R.D.S. and E.H.M.; investigation: R.D.S., L.M.D. and E.H.M.; data curation: R.D.S. and E.H.M.; writing—original draft: R.D.S. and E.H.M.; writing—review and editing: R.D.S., E.H.M., K.S.W. and R.G.J.; visualization: K.S.W.; supervision: R.G.J.; funding acquisition: R.G.J.
Corresponding author
Ethics declarations
Competing interests
R.G.J. is a consultant for Agios Pharmaceuticals and serves on the Scientific Advisory Board of ImmunoMet Therapeutics.
Additional information
Peer review information Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Ma, E. H. et al. Immunity 51, 856–870.e5 (2019): https://doi.org/10.1016/j.immuni.2019.09.003
Supplementary information
Supplementary Table 1
Compound list using the described HILIC LC/MS method. Important: Retention times will vary from lab to lab and should be confirmed by chemical standards.
Supplementary Table 2
Compound list using the described GC/MS method. The quantifier ion is a molecular ion that can be used for isotope analysis. The qualifier ion should have the same retention and is used to improve confidence in compound identification. Important: Retention times will vary from lab to lab and should be confirmed by chemical standards.
Supplementary Table 3
This Excel document contains three tabs. The “Raw Tracing Data” tab contains raw peak areas and natural abundance corrected values and fractional enrichment for each compound. The “T cells total 13C-enrichment” contains the total fractional enrichment of 13C carbon in each compound. These data have been filtered to include only compounds with at least 5% enrichment on average in at least one group. The “Subset >5% enrichment” contains this filtered data. In each tab, sample identifiers are as follows: Phenformin group: S1-00, S1-01, S1-30; Vehicle group: S2-03, S2-10, S2-30.
Rights and permissions
About this article
Cite this article
Sheldon, R.D., Ma, E.H., DeCamp, L.M. et al. Interrogating in vivo T-cell metabolism in mice using stable isotope labeling metabolomics and rapid cell sorting. Nat Protoc 16, 4494–4521 (2021). https://doi.org/10.1038/s41596-021-00586-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00586-2
This article is cited by
-
Principles of reproducible metabolite profiling of enriched lymphocytes in tumors and ascites from human ovarian cancer
Nature Protocols (2022)
-
Systems-based approaches to study immunometabolism
Cellular & Molecular Immunology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.