Abstract
The EGFR kinase pathway is one of the most frequently activated signaling pathways in human cancers. EGFR and HER2 are the two significant members of this pathway, which are attractive drug targets of clinical relevance in lung and breast cancer. Therefore, identifying EGFR- and HER2-specific inhibitors is one of the important challenges in cancer drug discovery. To address this issue, a dataset of 519 compounds having inhibitory activity against both the isoforms, i.e., EGFR and HER2, was collected from the literature and developed a knowledge-based computational classification model for predicting the specificity of a molecule for an isoform (EGFR/HER2) with precision. A total of seventy-two classification models using nine fingerprint types, four classifiers (IBK, NB, SMO and RF) and two different datasets (EGFR and HER2 isoform specific) were developed. It was observed that the models developed using random forest and IBK performed better for EGFR- and HER2-specific datasets, respectively. Scaffold and functional group analysis led to the identification of prevalent core and fragments in each of the datasets. The accuracy of the selected best performing models was also evaluated using the decoy dataset. We have also developed an application EGFRisopred, which integrates the best performing models and permits the user to predict the specificity of a compound as an EGFR-/HER2-specific anticancer agent. It is expected that the tool’s availability as a free utility will allow researchers to identify new inhibitors against these targets important in cancer.
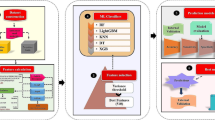
Graphic abstract






Similar content being viewed by others
References
Lemmon MA, Schlessinger J (2010) Cell signaling by receptor tyrosine kinases. Cell 141:1117–1134. https://doi.org/10.1016/j.cell.2010.06.011
Zwick E, Bange J, Ullrich A (2001) Receptor tyrosine kinase signalling as a target for cancer intervention strategies. Endocr Relat Cancer 8:161–173. https://doi.org/10.1677/erc.0.0080161
Baselga J, Swain SM (2009) Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer 9:463–475. https://doi.org/10.1038/nrc2656
De Castro DG, Clarke PA, Al-Lazikani B, Workman P (2013) Personalized cancer medicine: molecular diagnostics, predictive biomarkers, and drug resistance. Clin Pharmacol Ther 93:252–259. https://doi.org/10.1038/clpt.2012.237
Yadav IS, Nandekar PP, Shrivastava S et al (2014) Ensemble docking and molecular dynamics identify knoevenagel curcumin derivatives with potent anti-EGFR activity. Gene 539:82–90. https://doi.org/10.1016/j.gene.2014.01.056
Sharma VK, Nandekar PP, Sangamwar A et al (2016) Structure guided design and binding analysis of EGFR inhibiting analogues of erlotinib and AEE788 using ensemble docking, molecular dynamics and MM-GBSA. RSC Adv 6:65725–65735. https://doi.org/10.1039/c6ra08517b
Fatima S, Pal D, Agarwal SM (2019) QSAR of clinically important EGFR mutant L858R/T790M pyridinylimidazole inhibitors. Chem Biol Drug Des 94:1306–1315. https://doi.org/10.1111/cbdd.13505
Elmetwally SA, Saied KF, Eissa IH, Elkaeed EB (2019) Design, synthesis and anticancer evaluation of thieno[2,3-d]pyrimidine derivatives as dual EGFR/HER2 inhibitors and apoptosis inducers. Bioorg Chem 88:102944. https://doi.org/10.1016/j.bioorg.2019.102944
Gogoi D, Baruah VJ, Chaliha AK et al (2016) 3D pharmacophore-based virtual screening, docking and density functional theory approach towards the discovery of novel human epidermal growth factor receptor-2 (HER2) inhibitors. J Theor Biol 411:68–80. https://doi.org/10.1016/j.jtbi.2016.09.016
Issa NT, Stathias V, Schürer S, Dakshanamurthy S (2021) Machine and deep learning approaches for cancer drug repurposing. Semin Cancer Biol 68:132–142. https://doi.org/10.1016/j.semcancer.2019.12.011
Miljković F, Rodríguez-Pérez R, Bajorath J (2020) Machine learning models for accurate prediction of kinase inhibitors with different binding modes. J Med Chem 63:8738–8748. https://doi.org/10.1021/acs.jmedchem.9b00867
Dhiman K, Agarwal SM (2016) NPred: QSAR classification model for identifying plant based naturally occurring anti-cancerous inhibitors. RSC Adv 6:49395–49400. https://doi.org/10.1039/c6ra02772e
Saini R, Fatima S, Agarwal SM (2020) TMLRpred: A machine learning classification model to distinguish reversible EGFR double mutant inhibitors. Chem Biol Drug Des 96:921–930. https://doi.org/10.1111/cbdd.13697
Karpov P, Godin G, Tetko IV (2020) Transformer-CNN: Swiss knife for QSAR modeling and interpretation. J Cheminform 12:1–12. https://doi.org/10.1186/s13321-020-00423-w
Singh H, Singh S, Singla D et al (2015) QSAR based model for discriminating EGFR inhibitors and non-inhibitors using Random forest. Biol Direct 10:1–12. https://doi.org/10.1186/s13062-015-0046-9
Yadav I, Singh H, Khan M et al (2014) EGFRIndb: epidermal growth factor receptor inhibitor database. Anticancer Agents Med Chem 14:928–935. https://doi.org/10.2174/1871520614666140323203140
Tetko IV, Maran U, Tropsha A (2017) Public (Q)SAR services, integrated modeling environments, and model repositories on the web: state of the art and perspectives for future development. Mol Inform 36:1–14. https://doi.org/10.1002/minf.201600082
Cw YAP (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32:1466–1474
Danishuddin KA, Mobeen F, Khan AU (2018) Development of Ligand and Structure-based classification models to design novel inhibitors against antibiotic hydrolyzing enzymes: Integration of web server. J Biomol Struct Dyn 36:2966–2975. https://doi.org/10.1080/07391102.2017.1373034
Sander T, Freyss J, Von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55:460–473. https://doi.org/10.1021/ci500588j
Cao Y, Charisi A, Cheng LC et al (2008) ChemmineR: a compound mining framework for R. Bioinformatics 24:1733–1734. https://doi.org/10.1093/bioinformatics/btn307
Melagraki G, Afantitis A, Sarimveis H et al (2010) In silico exploration for identifying structure-activity relationship of MEK inhibition and oral bioavailability for isothiazole derivatives. Chem Biol Drug Des 76:397–406. https://doi.org/10.1111/j.1747-0285.2010.01029.x
Afantitis A, Melagraki G, Koutentis PA et al (2011) Ligand - Based virtual screening procedure for the prediction and the identification of novel β-amyloid aggregation inhibitors using Kohonen maps and counterpropagation artificial neural networks. Eur J Med Chem 46:497–508. https://doi.org/10.1016/j.ejmech.2010.11.029
Ishikawa T, Seto M, Banno H et al (2011) Design and synthesis of novel human epidermal growth factor receptor 2 (HER2)/epidermal growth factor receptor (EGFR) dual inhibitors bearing a pyrrolo[3,2-d]pyrimidine scaffold. J Med Chem 54:8030–8050. https://doi.org/10.1021/jm2008634
Ducray R, Ballard P, Barlaam BC et al (2008) Novel 3-alkoxy-1H-pyrazolo[3,4-d]pyrimidines as EGFR and erbB2 receptor tyrosine kinase inhibitors. Bioorganic Med Chem Lett 18:959–962. https://doi.org/10.1016/j.bmcl.2007.12.035
Stevens KL, Alligood KJ, Alberti JGB et al (2009) Synthesis and stereochemical effects of pyrrolidinyl-acetylenic thieno[3,2-d]pyrimidines as EGFR and ErbB-2 inhibitors. Bioorganic Med Chem Lett 19:21–26. https://doi.org/10.1016/j.bmcl.2008.11.023
Wissner A, Overbeek E, Reich MF et al (2003) Synthesis and structure-activity relationships of 6,7-disubstituted 4-anilinoquinoline-3-carbonitriles. The design of an orally active, irreversible inhibitor of the tyrosine kinase activity of the epidermal growth factor receptor (EGFR) and the human epi. J Med Chem 46:49–63. https://doi.org/10.1021/jm020241c
Denny WA (2001) The 4-anilinoquinazoline class of inhibitors of the erbB family of receptor tyrosine kinases. Farmaco 56:51–56. https://doi.org/10.1016/S0014-827X(01)01026-6
Yun CH, Boggon TJ, Li Y et al (2007) Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 11:217–227. https://doi.org/10.1016/j.ccr.2006.12.017
Pawar VG, Sos ML, Rode HB et al (2010) Synthesis and biological evaluation of 4-anilinoquinolines as potent inhibitors of epidermal growth factor receptor. J Med Chem 53:2892–2901. https://doi.org/10.1021/jm901877j
Waterson AG, Stevens KL, Reno MJ et al (2006) Alkynyl pyrimidines as dual EGFR/ErbB2 kinase inhibitors. Bioorganic Med Chem Lett 16:2419–2422. https://doi.org/10.1016/j.bmcl.2006.01.111
Gandin V, Ferrarese A, Dalla Via M et al (2015) Targeting kinases with anilinopyrimidines: discovery of N-phenyl-N’-[4-(pyrimidin-4-ylamino)phenyl]urea derivatives as selective inhibitors of class III receptor tyrosine kinase subfamily. Sci Rep 5:1–16. https://doi.org/10.1038/srep16750
Fink BE, Norris D, Mastalerz H et al (2011) Novel pyrrolo[2,1-f][1,2,4]triazin-4-amines: dual inhibitors of EGFR and HER2 protein tyrosine kinases. Bioorganic Med Chem Lett 21:781–785. https://doi.org/10.1016/j.bmcl.2010.11.100
Rheault TR, Caferro TR, Dickerson SH et al (2009) Thienopyrimidine-based dual EGFR/ErbB-2 inhibitors. Bioorganic Med Chem Lett 19:817–820. https://doi.org/10.1016/j.bmcl.2008.12.011
Kawakita Y, Banno H, Ohashi T et al (2012) Design and synthesis of pyrrolo[3,2- d ]pyrimidine human epidermal growth factor receptor 2 (HER2)/epidermal growth factor receptor (EGFR) dual inhibitors: Exploration of novel back-pocket binders. J Med Chem 55:3975–3991. https://doi.org/10.1021/jm300185p
Milik SN, Abdel-Aziz AK, Lasheen DS et al (2018) Surmounting the resistance against EGFR inhibitors through the development of thieno[2,3-d]pyrimidine-based dual EGFR/HER2 inhibitors. Eur J Med Chem 155:316–336. https://doi.org/10.1016/j.ejmech.2018.06.011
Ballard P, Bradbury RH, Hennequin LFA et al (2005) 5-Substituted 4-anilinoquinazolines as potent, selective and orally active inhibitors of erbB2 receptor tyrosine kinase. Bioorganic Med Chem Lett 15:4226–4229. https://doi.org/10.1016/j.bmcl.2005.06.068
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J Med Chem 55:6582–6594. https://doi.org/10.1021/jm300687e
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ravi Saini: Work done at ICMR-National Institute of Cancer Prevention and Research.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saini, R., Agarwal, S.M. EGFRisopred: a machine learning-based classification model for identifying isoform-specific inhibitors against EGFR and HER2. Mol Divers 26, 1531–1543 (2022). https://doi.org/10.1007/s11030-021-10284-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10284-6