Malpigmentation of Common Sole (Solea solea) during Metamorphosis Is Associated with Differential Synaptic-Related Gene Expression
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. RNA Extraction and Sequencing
2.3. Bioinformatics Analysis
2.3.1. Read Pre-Processing
2.3.2. Transcriptome Assembly and Annotation
3. Results
3.1. Transcriptome Assembly
3.2. Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAM | a disintegrin and metalloproteinase |
| AKT | protein kinase B |
| AMPA | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| ARA | arachidonic acid |
| ATP | adenosine triphosphate |
| CaMKII | calmodulin-dependent protein kinase II |
| CE | cornified envelope |
| CEGMA | core eukaryotic genes mapping approach |
| CREB | cyclic-AMP response element-binding protein |
| DHA | docosahexaenoic acid |
| dph | days post-hatching |
| EPA | eicosapentaenoic acid |
| EPTP | epitempin |
| FAK | focal adhesion kinase |
| FDR | Benjamini–Hochberg false discovery rate |
| FLG | filaggrin |
| GDP | guanosine 5′-diphosphate |
| GluR1,2,3,4 | glutamate receptor 1,2,3,4 |
| GRIP | glutamate receptor interacting protein |
| GTP | guanosine 5′-triphosphate |
| hpf | hours post fertilization |
| LGI | leucine-rich glioma inactivated |
| LTP | long term potentiation |
| MCH | melanin-concentrating hormone |
| NCBI | national center for biotechnology information |
| NGS | next-generation sequencing |
| NMDA | N-methyl-D-aspartate |
| NSF | N-ethylmaleimide-sensitive fusion protein |
| PERP | p53 effector related to PMP-22 |
| PICK | protein interacting with C kinase |
| PKC | protein kinase C |
| POMC | pro-opiomelanocortin |
| PSD95 | postsynaptic density protein 95 |
| SNAP | soluble NSF attachment protein |
| SNARE | soluble NSF attachment receptor |
| α-MSH | α-Melanocyte-stimulating hormone |
References
- Burton, D. The chromatic biology of flatfish (Pleuronectidae). Ital. J. Zool. 1998, 65, 399–403. [Google Scholar] [CrossRef]
- Washio, Y.; Aritaki, M.; Fujinami, Y.; Shimizu, D.; Yokoi, H.; Suzuki, T. Ocular-Side Lateralization of Adult-Type Chromatophore Precursors: Development of Pigment Asymmetry in Metamorphosing Flounder Larvae. J. Exp. Zool. Part B Mol. Dev. Evol. 2013, 320, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Bolker, J.A.; Hakala, T.F.; Quist, J.E. Pigmentation development, defects, and patterning in summer flounder (Paralichthys dentatus). Zoology 2005, 108, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Guillot, R.; Ceinos, R.M.; Cal, R.; Rotllant, J.; Cerdá-Reverter, J.M. Transient Ectopic Overexpression of Agouti-Signalling Protein 1 (Asip1) Induces Pigment Anomalies in Flatfish. PLoS ONE 2012, 7, e48526. [Google Scholar] [CrossRef] [Green Version]
- Bolker, J.A. Pigmentation development in hatchery-reared flatfishes. J. Fish Biol. 2000, 56, 1029–1052. [Google Scholar] [CrossRef]
- Venizelos, A.; Benetti, D.D. Pigment abnormalities in flatfish. Aquaculture 1999, 176, 181–188. [Google Scholar] [CrossRef]
- Yamanome, T.; Amano, M.; Takahashi, A. White background reduces the occurrence of staining, activates melanin-concentrating hormone and promotes somatic growth in barfin flounder. Aquaculture 2005, 244, 323–329. [Google Scholar] [CrossRef]
- Imsland, A.K.; Wergeland, T.; Jonassen, T.M.; Stefansson, S.O. Does malpigmentation improve growth in juvenile turbot (Scophthalmus maximus Rafinesque) and halibut (Hippoglossus hippoglossus L.)? Aquac. Res. 2006, 37, 306–312. [Google Scholar] [CrossRef]
- Lund, I.; Steenfeldt, S.J.; Hansen, B.W. Influence of dietary arachidonic acid combined with light intensity and tank colour on pigmentation of common sole (Solea solea L.) larvae. Aquaculture 2010, 308, 159–165. [Google Scholar] [CrossRef]
- Vizcaíno-Ochoa, V.; Lazo, J.P.; Barón-Sevilla, B.; Drawbridge, M.A. The effect of dietary docosahexaenoic acid (DHA) on growth, survival and pigmentation of California halibut Paralichthys californicus larvae (Ayres, 1810). Aquaculture 2010, 302, 228–234. [Google Scholar] [CrossRef]
- Isojima, T.; Tsuji, H.; Masuda, R.; Tagawa, M. Formation process of staining-type hypermelanosis in Japanese flounder juveniles revealed by examination of chromatophores and scales. Fish. Sci. 2013, 79, 231–242. [Google Scholar] [CrossRef] [Green Version]
- Akyol, O.; Şen, H. First record of abnormal pigmentation in a wild common sole, Solea solea L., from the Aegean Sea. Turkish J. Vet. Anim. Sci. 2012, 36, 727–729. [Google Scholar] [CrossRef]
- Conceição, L.E.C.; Ribeiro, L.; Engrola, S.; Aragão, C.; Morais, S.; Lacuisse, M.; Soares, F.; Dinis, M.T. Nutritional physiology during development of Senegalese sole (Solea senegalensis). Aquaculture 2007, 268, 64–81. [Google Scholar] [CrossRef]
- Exadactylos, A.; Malandrakis, E.E.; Panagiotaki, P.; Geffen, A.J. The development of size variation in Dover sole, Solea solea and turbot, Scophthalmus maximus: Genetic variability between different geographical and among year class farmed strains. Aquac. Res. 2013. [Google Scholar] [CrossRef]
- Kavouras, M.; Malandrakis, E.E.; Golomazou, E.; Konstantinidis, I.; Blom, E.; Palstra, A.P.; Anastassiadis, K.; Panagiotaki, P.; Exadactylos, A. Hox gene expression profiles during embryonic development of common sole. Anim. Biol. 2019. [Google Scholar] [CrossRef]
- Lund, I.; Steenfeldt, S.J.; Hansen, B.W. Effect of dietary arachidonic acid, eicosapentaenoic acid and docosahexaenoic acid on survival, growth and pigmentation in larvae of common sole (Solea solea L.). Aquaculture 2007, 273, 532–544. [Google Scholar] [CrossRef]
- Kang, D.Y.; Kim, H.C.; Kang, H.S. The functional relevance of prepro-melanin concentrating hormone (pMCH) to skin color change, Blind-side malpigmentation and feeding of oliver flounder Paralichthys olivaceus. Fish. Aquat. Sci. 2014, 17, 325–337. [Google Scholar] [CrossRef]
- Boglino, A.; Wishkerman, A.; Darias, M.J.; de la Iglesia, P.; Estévez, A.; Andree, K.B.; Gisbert, E. The effects of dietary arachidonic acid on Senegalese sole morphogenesis: A synthesis of recent findings. Aquaculture 2014, 432, 443–452. [Google Scholar] [CrossRef]
- Boglino, A.; Wishkerman, A.; Darias, M.J.; de la Iglesia, P.; Andree, K.B.; Gisbert, E.; Estévez, A. Senegalese sole (Solea senegalensis) metamorphic larvae are more sensitive to pseudo-albinism induced by high dietary arachidonic acid levels than post-metamorphic larvae. Aquaculture 2014, 433, 276–287. [Google Scholar] [CrossRef]
- Yamada, T.; Donai, H.; Okauchi, M.; Tagawa, M.; Araki, K. Induction of ambicoloration by exogenous cortisol during metamorphosis of spotted halibut Verasper variegatus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 160, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Cal, L.; Suarez-Bregua, P.; Moran, P.; Cerdá-Reverter, J.M.; Rotllant, J. Fish pigmentation. A key issue for the sustainable development of fish farming. In Emerging Issues in Fish Larvae Research; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Buzek, J.; Chastel, O. Directive 2010/63/EU of the European Parliament and of the Counsil of 22 September 2010 on the protection of animals used for scientific purposes. Off. J. Eur. Union 2010, OJ L 276, 33–79. [Google Scholar]
- Blonk, R.J.W.; Komen, J.; Tenghe, A.; Kamstra, A.; van Arendonk, J.A.M. Heritability of shape in common sole, Solea solea, estimated from image analysis data. Aquaculture 2010, 307, 6–11. [Google Scholar] [CrossRef]
- Kavouras, M.; Malandrakis, E.E.; Danis, T.; Blom, E.; Anastassiadis, K.; Panagiotaki, P.; Exadactylos, A. Hox genes polymorphism depicts developmental disruption of common sole eggs. Open Life Sci. 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Exadactylos, A.; Rigby, M.J.; Geffen, A.J.; Thorpe, J.P. Conservation aspects of natural populations and captive-bredstocks of turbot (Scophthalmus maximus) and Dover sole (Solea solea) using estimates of genetic diversity. ICES J. Mar. Sci. 2007, 64, 1173–1181. [Google Scholar] [CrossRef] [Green Version]
- Exadactylos, A.; Geffen, A.J.; Thorpe, J.P. Growth and genetic variation in hatchery-reared larval and juvenile Dover sole, Solea solea (L.). Aquaculture 1999, 176, 209–226. [Google Scholar] [CrossRef]
- Exadactylos, A.; Geffen, A.J.; Thorpe, J.P. Population structure of the Dover sole, Solea solea L., in a background of high gene flow. J. Sea Res. 1998, 40, 117–129. [Google Scholar] [CrossRef]
- Andrews, S. Fastqc: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Parra, G.; Bradnam, K.; Korf, I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007, 23, 1061–1067. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef] [Green Version]
- Sheng, M. Postsynaptic Signaling and Plasticity Mechanisms. Science 2002, 298, 776–780. [Google Scholar] [CrossRef]
- Bellono, N.W.; Oancea, E.V. Ion transport in pigmentation. Arch. Biochem. Biophys. 2014, 563. [Google Scholar] [CrossRef] [Green Version]
- Cull-Candy, S.; Kelly, L.; Farrant, M. Regulation of Ca2+-permeable AMPA receptors: Synaptic plasticity and beyond. Curr. Opin. Neurobiol. 2006, 16, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K. Synaptic plasticity and phosphorylation. Pharmacol. Ther. 2006, 112, 810–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kessels, H.W.; Malinow, R. Synaptic AMPA Receptor Plasticity and Behavior. Neuron 2009, 61, 340–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogundele, O.M.; Okunnuga, A.A.; Fabiyi, T.D.; Olajide, O.J.; Akinrinade, I.D.; Adeniyi, P.A.; Ojo, A.A. NMDA-R inhibition affects cellular process formation in Tilapia Melanocytes; a model for pigmented adrenergic neurons in process formation and retraction. Metab. Brain Dis. 2014, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Cull-Candy, S.G. Subunit interaction with PICK and GRIP controls Ca2+ permeability of AMPARs at cerebellar synapses. Nat. Neurosci. 2005, 8, 768–775. [Google Scholar] [CrossRef]
- Kropf, M.; Rey, G.; Glauser, L.; Kulangara, K.; Johnsson, K.; Hirling, H. Subunit-specific surface mobility of differentially labeled AMPA receptor subunits. Eur. J. Cell Biol. 2008, 87, 763–778. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Ziff, E.B. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron 2005, 47, 407–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, E. Molecular mechanism of neuronal plasticity: Induction and maintenance of long-term potentiation in the hippocampus. J. Pharmacol. Sci. 2006, 100, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Krapivinsky, G.; Krapivinsky, L.; Manasian, Y.; Ivanov, A.; Tyzio, R.; Pellegrino, C.; Ben-Ari, Y.; Clapham, D.E.; Medina, I. The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron 2003, 40, 775–784. [Google Scholar] [CrossRef] [Green Version]
- Djurdjevič, I.; Furmanek, T.; Miyazawa, S.; Sušnik Bajec, S. Comparative transcriptome analysis of trout skin pigment cells. BMC Genom. 2019, 20, 359. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; McMurray, C.T. Calmodulin kinase II attenuation of gene transcription by preventing cAMP response element-binding protein (CREB) dimerization and binding of the creb-binding protein. J. Biol. Chem. 2001, 276, 1735–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kegel, L.; Aunin, E.; Meijer, D.; Bermingham, J.R. LGI proteins in the nervous system. ASN Neuro 2013, 5, 167–181. [Google Scholar] [CrossRef]
- Gu, Q. Neuromodulatory transmitter systems in the cortex and their role in cortical plasticity. Neuroscience 2002, 111, 815–835. [Google Scholar] [CrossRef]
- Novak, U. ADAM proteins in the brain. J. Clin. Neurosci. 2004, 11, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Seals, D.F.; Courtneidge, S.A. The ADAMs family of metalloproteases: Multidomain proteins with multiple functions. Genes Dev. 2003, 17, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.-J.; Zhou, L.; Jiang, N.; Zhang, N.; Zou, N.; Zhou, L.; Wang, Y.; Cowell, J.K.; Shen, Y. Essential roles of leucine-rich glioma inactivated 1 in the development of embryonic and postnatal cerebellum. Sci. Rep. 2015, 5, 7827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukata, Y.; Adesnik, H.; Iwanaga, T.; Bredt, D.S.; Nicoll, R.A.; Fukata, M. Epilepsy-Related Ligand/Receptor Complex LGI1 and ADAM22 Regulate Synaptic Transmission. Science 2006, 313, 1792–1795. [Google Scholar] [CrossRef] [PubMed]
- Fukata, Y.; Lovero, K.L.; Iwanaga, T.; Watanabe, A.; Yokoi, N.; Tabuchi, K.; Shigemoto, R.; Nicoll, R.A.; Fukata, M. Disruption of LGI1-linked synaptic complex causes abnormal synaptic transmission and epilepsy. Proc. Natl. Acad. Sci. USA 2010, 107, 3799–3804. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, E.; Sagane, K.; Nagasu, T.; Kuromitsu, J. Altered nociceptive response in ADAM11-deficient mice. Brain Res. 2006, 1097, 39–42. [Google Scholar] [CrossRef]
- Takahashi, E.; Sagane, K.; Oki, T.; Yamazaki, K.; Nagasu, T.; Kuromitsu, J. Deficits in spatial learning and motor coordination in ADAM11-deficient mice. BMC Neurosci. 2006, 7, 19. [Google Scholar] [CrossRef] [Green Version]
- Abe, T.; Kudo, H. Molecular characterization and gene expression of syntaxin-1 and VAMP2 in the olfactory organ and brain during both seaward and homeward migrations of chum salmon, Oncorhynchus keta. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2019, 227. [Google Scholar] [CrossRef]
- Park, W.J.; Lim, Y.Y.; Kwon, N.S.; Baek, K.J.; Kim, D.S.; Yun, H.Y. Leucine-rich glioma inactivated 3 induces neurite outgrowth through akt and focal adhesion kinase. Neurochem. Res. 2010, 35, 789–796. [Google Scholar] [CrossRef]
- Kimura, N.; Ishii, Y.; Suzaki, S.; Negishi, T.; Kyuwa, S.; Yoshikawa, Y. Aβ upregulates and colocalizes with LGI3 in cultured rat astrocytes. Cell. Mol. Neurobiol. 2007, 27, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Okabayashi, S.; Kimura, N. LGI3 interacts with flotillin-1 to mediate APP trafficking and exosome formation. Neuroreport 2010, 21, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Okabayashi, S.; Kimura, N. Leucine-rich glioma inactivated 3 is involved in amyloid beta peptide uptake by astrocytes and endocytosis itself. Neuroreport 2008, 19, 1175–1179. [Google Scholar] [CrossRef] [PubMed]
- Kühlbrandt, W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004, 5, 282–295. [Google Scholar] [CrossRef]
- Cokus, S.J.; De La Torre, M.; Medina, E.F.; Rasmussen, J.P.; Ramirez-Gutierrez, J.; Sagasti, A.; Wang, F. Tissue-Specific Transcriptomes Reveal Gene Expression Trajectories in Two Maturing Skin Epithelial Layers in Zebrafish Embryos. G3 Genes Genomes Genet. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, E. Keratins and the Skin. Annu. Rev. Cell Dev. Biol. 1995, 11, 123–154. [Google Scholar] [CrossRef]
- Waschke, J. The desmosome and pemphigus. Histochem. Cell Biol. 2008, 130, 21–54. [Google Scholar] [CrossRef] [Green Version]
- Moll, R.; Divo, M.; Langbein, L. The human keratins: Biology and pathology. Histochem. Cell Biol. 2008, 129, 705–733. [Google Scholar] [CrossRef] [Green Version]
- Mack, J.W.; Steven, A.C.; Steinert, P.M. The mechanism of interaction of filaggrin with intermediate filaments. The ionic zipper hypothesis. J. Mol. Biol. 1993, 232, 50–66. [Google Scholar] [CrossRef]
- Gruber, R.; Elias, P.M.; Crumrine, D.; Lin, T.K.; Brandner, J.M.; Hachem, J.P.; Presland, R.B.; Fleckman, P.; Janecke, A.R.; Sandilands, A.; et al. Filaggrin genotype in ichthyosis vulgaris predicts abnormalities in epidermal structure and function. Am. J. Pathol. 2011, 178, 2252–2263. [Google Scholar] [CrossRef] [Green Version]
- Candi, E.; Schmidt, R.; Melino, G. The cornified envelope: A model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 2005, 6, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Attardi, L.D.; Reczek, E.E.; Cosmas, C.; Demicco, E.G.; McCurrach, M.E.; Lowe, S.W.; Jacks, T. PERP, an apoptosis-associated target of p53, in a novel member of the PMP-22/gas3 family. Genes Dev. 2000, 14, 704–718. [Google Scholar] [CrossRef] [PubMed]
- Ihrie, R.A.; Marques, M.R.; Nguyen, B.T.; Horner, J.S.; Papazoglu, C.; Bronson, R.T.; Mills, A.A.; Attardi, L.D. Perp is a p63-regulated gene essential for epithelial integrity. Cell 2005, 120, 843–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martsikalis, P.V.; Gkafas, G.A.; Palaiokostas, C.; Exadactylos, A. Genomics Era on Breeding Aquaculture Stocks. In Organic Aquaculture; Springer: Cham, Switzerland, 2019. [Google Scholar]



| Pathway Identifier | Pathway Name | FDR | |
|---|---|---|---|
| 1 | R-DRE-438066 | Unblocking of NMDA receptor, glutamate binding, and activation | 2.81 × 10−11 |
| 2 | R-DRE-936837 | Ion transport by P-type ATPases | 3.47 × 10−10 |
| 3 | R-DRE-6805567 | Keratinization | 3.97 × 10−10 |
| 4 | R-DRE-6809371 | Formation of the cornified envelope | 3.97 × 10−10 |
| 5 | R-DRE-446107 | Type I hemidesmosome assembly | 4.83 × 10−9 |
| 6 | R-DRE-5578775 | Ion homeostasis | 2.09 × 10−8 |
| 7 | R-DRE-442755 | Activation of NMDA receptor and postsynaptic events | 4.01 × 10−8 |
| 8 | R-DRE-399710 | Activation of AMPA receptors | 8.50 × 10−7 |
| 9 | R-DRE-112314 | Neurotransmitter receptors and postsynaptic signal transmission | 8.50 × 10−7 |
| 10 | R-DRE-416993 | Trafficking of GluR2-containing AMPA receptors | 3.49 × 10−6 |
| 11 | R-DRE-399719 | Trafficking of AMPA receptors | 3.49 × 10−6 |
| 12 | R-DRE-399721 | Glutamate binding, activation of AMPA receptors, and synaptic plasticity | 3.49 × 10−6 |
| 13 | R-DRE-983712 | Ion channel transport | 3.86 × 10−6 |
| 14 | R-DRE-112315 | Transmission across Chemical Synapses | 5.62 × 10−6 |
| 15 | R-DRE-5576891 | Cardiac conduction | 2.24 × 10−5 |
| 16 | R-DRE-442729 | CREB phosphorylation through the activation of CaMKII | 2.70 × 10−5 |
| 17 | R-DRE-442982 | Ras activation upon Ca2+ infux through NMDA receptor | 3.53 × 10−5 |
| 18 | R-DRE-397014 | Muscle contraction | 8.81 × 10−5 |
| 19 | R-DRE-1266738 | Developmental Biology | 8.81 × 10−5 |
| 20 | R-DRE-5682910 | LGI-ADAM interactions | 1.02 × 10−4 |
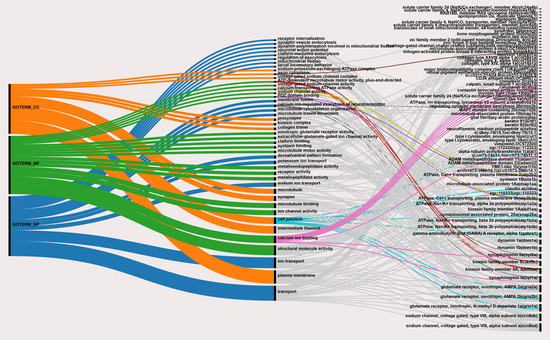
| Category | Term | Fisher Exact |
|---|---|---|
| GO_BIOLOGICAL_PROCESS | ion transport | 2.90 × 10−6 |
| sodium ion transport | 8.70 × 10−5 | |
| receptor internalization | 1.30 × 10−4 | |
| synaptic vesicle endocytosis | 2.60 × 10−4 | |
| transport | 3.70 × 10−4 | |
| dynamin polymerization involved in mitochondrial fission | 6.40 × 10−4 | |
| membrane fusion | 9.10 × 10−4 | |
| neuronal action potential | 1.50 × 10−3 | |
| clathrin-mediated endocytosis | 1.90 × 10−3 | |
| calcium ion-regulated exocytosis of neurotransmitter | 2.70 × 10−3 | |
| regulation of exocytosis | 3.80 × 10−3 | |
| mitochondrial fission | 4.30 × 10−3 | |
| larval locomotory behavior | 4.30 × 10−3 | |
| dorsal/ventral pattern formation | 6.20 × 10−3 | |
| potassium ion transport | 7.60 × 10−3 | |
| microtubule cytoskeleton organization | 8.10 × 10−3 | |
| microtubule-based movement | 1.60 × 10−2 | |
| GO_CELLULAR_COMPONENT | intermediate filament | 6.20 × 10−10 |
| microtubule | 2.50 × 10−4 | |
| cell junction | 2.50 × 10−4 | |
| synapse | 8.10 × 10−4 | |
| sodium:potassium-exchanging ATPase complex | 1.10 × 10−3 | |
| axon cytoplasm | 1.80 × 10−3 | |
| voltage-gated sodium channel complex | 3.90 × 10−3 | |
| presynapse | 6.10 × 10−3 | |
| kinesin complex | 9.10 × 10−3 | |
| collagen trimer | 1.30 × 10−2 | |
| plasma membrane | 2.40 × 10−2 | |
| GO_MOLECULAR_FUNCTION | structural molecule activity | 5.40 × 10−10 |
| microtubule binding | 3.80 × 10−4 | |
| ionotropic glutamate receptor activity | 7.40 × 10−4 | |
| extracellular-glutamate-gated ion channel activity | 8.30 × 10−4 | |
| ATP-dependent microtubule motor activity, plus-end-directed | 2.80 × 10−3 | |
| voltage-gated sodium channel activity | 2.80 × 10−3 | |
| calcium-transporting ATPase activity | 3.30 × 10−3 | |
| ion channel activity | 3.30 × 10−3 | |
| clathrin binding | 3.50 × 10−3 | |
| sodium channel activity | 3.80 × 10−3 | |
| PDZ domain binding | 4.30 × 10−3 | |
| metalloendopeptidase activity | 6.80 × 10−3 | |
| receptor activity | 8.90 × 10−3 | |
| syntaxin binding | 1.00 × 10−2 | |
| metallopeptidase activity | 1.10 × 10−2 | |
| microtubule motor activity | 1.50 × 10−2 | |
| calcium ion binding | 4.80 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kavouras, M.; Malandrakis, E.E.; Blom, E.; Tsilika, K.; Danis, T.; Panagiotaki, P.; Exadactylos, A. Malpigmentation of Common Sole (Solea solea) during Metamorphosis Is Associated with Differential Synaptic-Related Gene Expression. Animals 2021, 11, 2273. https://doi.org/10.3390/ani11082273
Kavouras M, Malandrakis EE, Blom E, Tsilika K, Danis T, Panagiotaki P, Exadactylos A. Malpigmentation of Common Sole (Solea solea) during Metamorphosis Is Associated with Differential Synaptic-Related Gene Expression. Animals. 2021; 11(8):2273. https://doi.org/10.3390/ani11082273
Chicago/Turabian StyleKavouras, Menelaos, Emmanouil E. Malandrakis, Ewout Blom, Kyriaki Tsilika, Theodoros Danis, Panagiota Panagiotaki, and Athanasios Exadactylos. 2021. "Malpigmentation of Common Sole (Solea solea) during Metamorphosis Is Associated with Differential Synaptic-Related Gene Expression" Animals 11, no. 8: 2273. https://doi.org/10.3390/ani11082273