Abstract
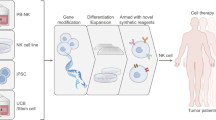
Efficacious and accessible sources of natural killer (NK) cells would widen their use as immunotherapeutics, particularly for solid cancers. Here, we show that human somatic cells can be directly reprogrammed into NK cells with a CD56brightCD16bright phenotype using pluripotency transcription factors and an optimized reprogramming medium. The directly reprogrammed NK cells have strong innate–adaptive immunomodulatory activity and are highly potent against a wide range of cancer cells, including difficult-to-treat solid cancers and cancer stem cells. Both directly reprogrammed NK cells bearing a cancer-specific chimeric antigen receptor and reprogrammed NK cells in combination with antibodies competent for antibody-dependent cell-mediated cytotoxicity led to selective anticancer effects with augmented potency. The direct reprogramming of human somatic cells into NK cells is amenable to the production of autologous and allogeneic NK cells, and will facilitate the design and testing of cancer immunotherapies and combination therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. All data generated in this study, including source data and the data used to make the figures, are available from Figshare at https://figshare.com/s/93edb79d917acacdcd20. The microarray data are available from the Gene Expression Omnibus under accession number GSE132907. Source data are provided with this paper.
References
Shimasaki, N., Jain, A. & Campana, D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 19, 200–218 (2020).
Chiossone, L., Dumas, P. Y., Vienne, M. & Vivier, E. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 18, 671–688 (2018).
Guillerey, C., Huntington, N. D. & Smyth, M. J. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 17, 1025–1036 (2016).
Caligiuri, M. A. Human natural killer cells. Blood 112, 461–469 (2008).
Tallerico, R. et al. Human NK cells selective targeting of colon cancer-initiating cells: a role for natural cytotoxicity receptors and MHC class I molecules. J. Immunol. 190, 2381–2390 (2013).
Messaoudene, M. et al. Mature cytotoxic CD56bright/CD16+ natural killer cells can infiltrate lymph nodes adjacent to metastatic melanoma. Cancer Res. 74, 81–92 (2014).
Cooper, M. A., Fehniger, T. A. & Caligiuri, M. A. The biology of human natural killer-cell subsets. Trends Immunol. 22, 633–640 (2001).
Verneris, M. R. & Miller, J. S. The phenotypic and functional characteristics of umbilical cord blood and peripheral blood natural killer cells. Br. J. Haematol. 147, 185–191 (2009).
Freud, A. G., Mundy-Bosse, B. L., Yu, J. & Caligiuri, M. A. The broad spectrum of human natural killer cell diversity. Immunity 47, 820–833 (2017).
Shereck, E. et al. Immunophenotypic, cytotoxic, proteomic and genomic characterization of human cord blood vs. peripheral blood CD56Dim NK cells. Innate Immun. 25, 294–304 (2019).
Jacobs, R. et al. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur. J. Immunol. 31, 3121–3127 (2001).
Jonges, L. E. et al. The phenotypic heterogeneity of human natural killer cells: presence of at least 48 different subsets in the peripheral blood. Scand. J. Immunol. 53, 103–110 (2001).
Marcon, F. et al. NK cells in pancreatic cancer demonstrate impaired cytotoxicity and a regulatory IL-10 phenotype. OncoImmunology 9, 1845424 (2020).
Alter, G. et al. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood 106, 3366–3369 (2005).
Silla, L. Double-bright (CD56bright/CD16bright) natural killer cell adoptive immunotherapy for SARS-CoV-2. Br. J. Haematol. 190, e322–e323 (2020).
Liu, E. et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 382, 545–553 (2020).
Tang, X. et al. First-in-man clinical trial of CAR NK-92 cells: safety test of CD33-CAR NK-92 cells in patients with relapsed and refractory acute myeloid leukemia. Am. J. Cancer Res. 8, 1083–1089 (2018).
Li, Y., Hermanson, D. L., Moriarity, B. S. & Kaufman, D. S. Human iPSC-derived natural killer cells engineered with chimeric antigen receptors enhance anti-tumor activity. Cell Stem Cell 23, 181–192.e5 (2018).
Zhu, H., Lai, Y. S., Li, Y., Blum, R. H. & Kaufman, D. S. Concise review: human pluripotent stem cells to produce cell-based cancer immunotherapy. Stem Cells 36, 134–145 (2018).
Zhu, H. et al. Pluripotent stem cell-derived NK cells with high-affinity noncleavable CD16a mediate improved antitumor activity. Blood 135, 399–410 (2020).
Rosa, F. F. et al. Direct reprogramming of fibroblasts into antigen-presenting dendritic cells. Sci. Immunol. 3, eaau4292 (2018).
Xie, H., Ye, M., Feng, R. & Graf, T. Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676 (2004).
Szabo, E. et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468, 521–526 (2010).
Galat, Y. et al. Application of small molecule CHIR99021 leads to the loss of hemangioblast progenitor and increased hematopoiesis of human pluripotent stem cells. Exp. Hematol. 65, 38–48.e1 (2018).
Cao, N. et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352, 1216–1220 (2016).
Roeven, M. W. et al. The aryl hydrocarbon receptor antagonist StemRegenin1 improves in vitro generation of highly functional natural killer cells from CD34+ hematopoietic stem and progenitor cells. Stem Cells Dev. 24, 2886–2898 (2015).
Angelos, M. G. et al. Aryl hydrocarbon receptor inhibition promotes hematolymphoid development from human pluripotent stem cells. Blood 129, 3428–3439 (2017).
Hughes, T. et al. The transcription factor AHR prevents the differentiation of a stage 3 innate lymphoid cell subset to natural killer cells. Cell Rep. 8, 150–162 (2014).
Ko, C. I. et al. Repression of the aryl hydrocarbon receptor is required to maintain mitotic progression and prevent loss of pluripotency of embryonic stem cells. Stem Cells 34, 2825–2839 (2016).
Ko, C. I., Wang, Q., Fan, Y., Xia, Y. & Puga, A. Pluripotency factors and Polycomb group proteins repress aryl hydrocarbon receptor expression in murine embryonic stem cells. Stem Cell Res. 12, 296–308 (2014).
Pegram, H. J., Andrews, D. M., Smyth, M. J., Darcy, P. K. & Kershaw, M. H. Activating and inhibitory receptors of natural killer cells. Immunol. Cell Biol. 89, 216–224 (2011).
Chen, X. et al. High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci. Rep. 4, 7481 (2014).
Ohtani, H. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. 7, 4 (2007).
Petriello, A. et al. Assessment of human natural killer cell events driven by FcγRIIIa engagement in the presence of therapeutic antibodies. J. Vis. Exp. https://doi.org/10.3791/61144 (2020).
Liu, S. D. et al. Afucosylated antibodies increase activation of FcγRIIIa-dependent signaling components to intensify processes promoting ADCC. Cancer Immunol. Res. 3, 173–183 (2015).
Jonker, D. J. et al. Cetuximab for the treatment of colorectal cancer. N. Engl. J. Med. 357, 2040–2048 (2007).
Ishikawa, T. et al. Phase I clinical trial of adoptive transfer of expanded natural killer cells in combination with IgG1 antibody in patients with gastric or colorectal cancer. Int. J. Cancer 142, 2599–2609 (2018).
Romee, R. et al. Cytokine activation induces human memory-like NK cells. Blood 120, 4751–4760 (2012).
Cerwenka, A. & Lanier, L. L. Natural killer cell memory in infection, inflammation and cancer. Nat. Rev. Immunol. 16, 112–123 (2016).
Bar-Nur, O. et al. Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat. Biotechnol. 33, 761–768 (2015).
Zhu, S., Wang, H. & Ding, S. Reprogramming fibroblasts toward cardiomyocytes, neural stem cells and hepatocytes by cell activation and signaling-directed lineage conversion. Nat. Protoc. 10, 959–973 (2015).
Hermanson, D. L. et al. Induced pluripotent stem cell-derived natural killer cells for treatment of ovarian cancer. Stem Cells 34, 93–101 (2016).
Zhu, H. & Kaufman, D. S. An improved method to produce clinical-scale natural killer cells from human pluripotent stem cells. Methods Mol. Biol. 2048, 107–119 (2019).
Kim, J. et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl Acad. Sci. USA 108, 7838–7843 (2011).
Maza, I. et al. Transient acquisition of pluripotency during somatic cell transdifferentiation with iPSC reprogramming factors. Nat. Biotechnol. 33, 769–774 (2015).
Grossenbacher, S. K., Canter, R. J. & Murphy, W. J. Natural killer cell immunotherapy to target stem-like tumor cells. J. Immunother. Cancer 4, 19 (2016).
Sandel, M. H. et al. Natural killer cells infiltrating colorectal cancer and MHC class I expression. Mol. Immunol. 42, 541–546 (2005).
Mace, E. M. Phosphoinositide-3-kinase signaling in human natural killer cells: new insights from primary immunodeficiency. Front. Immunol. 9, 445 (2018).
Klingemann, H., Boissel, L. & Toneguzzo, F. Natural killer cells for immunotherapy—advantages of the NK-92 cell line over blood NK cells. Front. Immunol. 7, 91 (2016).
Uppendahl, L. D. et al. Cytokine-induced memory-like natural killer cells have enhanced function, proliferation, and in vivo expansion against ovarian cancer cells. Gynecol. Oncol. 153, 149–157 (2019).
Vivier, E. et al. Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–49 (2011).
Ray, A. & Dittel, B. N. et al.Isolation of mouse peritoneal cavity cells. J. Vis. Exp. 2010, 1488 (2010).
Acknowledgements
We are grateful to I. Choi for insightful advice during the course of this investigation. This work was supported by the National Research Foundation of Korea (2020R1A2B5B02002252 and 2019M3A9H1103797), the National Research Council of Science and Technology (CRC-15-02-KRIBB) and the KRIBB Research Initiative Program (1711134084/KGM5502113).
Author information
Authors and Affiliations
Contributions
H.-S.K. and Y.S.C. conceived of the study idea. H.-S.K., B.S. and J.Y.K. developed the methodology. H.-S.K., B.S., J.Y.K., C.L.S., J.E.J. and Y.S.C. performed the investigation. H.-S.K., B.S. and J.Y.K. performed the statistical analyses. Y.S.C. provided resources. H.-S.K. and Y.S.C. wrote the original draft of the manuscript. H.-S.K. and Y.S.C. reviewed and edited the manuscript. Y.S.C. acquired funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biomedical Engineering thanks Pin Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Gene-expression profile in the reprogramming procedure.
a-c, RT-qPCR analysis of hematopoietic progenitor-related genes [C-KIT(CD117), CD27, CD34, CD38, CD49, and CD90] (a), NK-related genes (CD16, CD56, NCR1/NKP46, and NCR3/NKP30) (b), and T cell-related genes (CD3E, GATA3, HES1, and TCF1) (c) at the indicated days of reprogramming from CD3+ T cells. Mean ± SD (n = 3). Two-tailed Student’s t-test. *P < 0.001 (vs the starting cells, day 0).
Extended Data Fig. 2 Comparative cytolytic and cytokine-secretion activities of iNK cells, PBMC-NK cells and UCB-NK cells.
a, The cytolytic activity of the drNK cells, PBMC-NK cells, and UCB-NK cells against K562 blood cancer cells at the indicated E:T ratios. b, The cytolytic activity of the drNK cells, PBMC-NK cells, and UCB-NK cells against HepG2 liver cancer cells at the indicated E:T ratios. c, The cytokine (IFNγ, granzyme B, and TNF-α) secretion by drNK cells, PBMC-NK cells, and UCB-NK cells stimulated with or without cancer cells (K562 and HepG2) for 16 h at an E:T ratio of 1:1. Mean ± SD (n = 3 biological replicates for each sample). Two-tailed Student’s t-test. *P < 0.001 (vs no target cells).
Extended Data Fig. 3 T cells recruited by iNK cells invade solid tumours in vivo.
a, b, In T cell recruitment test in vivo (Fig. 6i), RT-qPCR analysis of T cell-specific genes (CD3D) (a), chemokines (CCL3, CCL4, and CCL5), and the cytokine IFN-γ in tumor specimens at day 8 (b). Mean ± SD (n = 3). Two-tailed Student’s t-test. *P < 0.001 (vs control).
Extended Data Fig. 4 Anticancer effects of iNK cells in the xenograft model.
a, Quantitative graph of the results shown in Fig. 8b. Quantification of bioluminescence signals at the indicated days (c). b, Representative images of tumors from the mice injected with PBS control, drNK-low (5.0 × 106), drNK cells (1.5 ×107), and doxorubicin (2 mg/kg) on day 28 post SW620-Luc inoculation. c, Anticancer effects of the indicated treatments were observed by assessing tumor volume in (b). Each symbol represents the tumor volume from the mouse in each group. Horizontal bars show the mean values, and error bars show the SD (n = 12 biological replicates for each sample). Two-tailed Student’s t-test. *P < 0.001 (vs no drNK cells). d, Infiltration of drNK cells in tumor xenografts, analyzed through both bioluminescence (tumor xenograft) and biofluorescence (DiR-drNK cells) 5 days after injection of drNK cells or PBS control.
Extended Data Fig. 5 Anticancer effects of CAR-iNK cells in the xenograft model.
a, b, Quantitative graph of the results shown in Fig. 8g. Quantification of bioluminescence signals at the indicated days (a) and on day 21 (b). Horizontal bars show the mean values, and error bars show the SD. Each symbol represents one tumor xenograft in each group. Mean ± SD (n = 8-10 biological replicates for each sample). Two-tailed Student’s t-test. *P < 0.001 (vs the control).c, Schematic of the in vivo anticancer activity assay of HER2-drNK cells. Mice bearing luciferase-expressing COLO 205 (COLO 205-Luc) xenografts were i.v. injected with either PBS, drNK cells, HER2-drNK cells, NK92 or HER2 CAR expressing NK-92 (HER2-NK92) on day 1. d, Representative bioluminescence images of the mice receiving the indicated treatments at the indicated days after COLO 205-Luc inoculation. Mean ± SD (n = 12 biological replicates for each sample). Mice bearing luciferase-expressing COLO 205 (COLO 205-Luc) xenografts were i.v. injected with either PBS, drNK cells, HER2-NK cells, NK-92 or HER2 CAR expressing NK-92 (HER2-NK-92) on day 1. e, f, Quantitative graph of the results shown in (d). Quantification of bioluminescence signals at the indicated days (e) and on day 21 (f). Horizontal bars show the mean values, and error bars show the SD. Each symbol represents one tumor xenograft in each group. Mean ± SD (n = 10-20 biological replicates for each sample). Two-tailed Student’s t-test. *P < 0.001 (vs the control).
Extended Data Fig. 6 T cells recruited by iNK cells promote iNK-cell antitumour effects.
a, Representative bioluminescence images of the mice receiving the indicated treatments at the indicated days after SW620-Luc inoculation. Mean ± SD (n = 6-12). Experimental design of in vivo anticancer activity assays of T cells recruited by drNK cells. The mice s.c. injected with luciferase-expressing SW620 cells (SW620-Luc) were i.v. injected with PBS or 1×107 drNK cells, activated T cells, or drNK cells and 5×106 activated T cells (n = 6 per group) on day 4. b, c, Quantitative graph of the results shown in (a). Quantification of bioluminescence signals at the indicated days (b) and on day 21 (c). Horizontal bars show the mean values, and error bars show the SD. Each symbol represents one mouse in each group. Mean ± SD (n = 6-8 biological replicates for each sample). Two-tailed Student’s t-test. *P < 0.001 (vs control).
Supplementary information
Supplementary Information
Supplementary Methods, Figs. 1–7, Tables 1 and 2 and References.
Source data
Source Data Fig. 1
Source data and statistics.
Source Data Fig. 2
Source data and statistics.
Source Data Fig. 3
Source data and statistics.
Source Data Fig. 4
Source data and statistics.
Source Data Fig. 5
Source data and statistics.
Source Data Fig. 6
Source data and statistics.
Source Data Fig. 7
Source data and statistics.
Source Data Fig. 8
Source data and statistics.
Extended DataSource Data Fig. 1
Source data and statistics.
Source Data Extended Data Fig. 2
Source data and statistics.
Source Data Extended Data Fig. 3
Source data and statistics.
Source Data Extended Data Fig. 4
Source data and statistics.
Source Data Extended Data Fig. 5
Source data and statistics.
Source Data Extended Data Fig. 6
Source data and statistics.
Rights and permissions
About this article
Cite this article
Kim, HS., Kim, J.Y., Seol, B. et al. Directly reprogrammed natural killer cells for cancer immunotherapy. Nat Biomed Eng 5, 1360–1376 (2021). https://doi.org/10.1038/s41551-021-00768-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00768-z
This article is cited by
-
Prospects for Development of Induced Pluripotent Stem Cell-Derived CAR-Targeted Immunotherapies
Archivum Immunologiae et Therapiae Experimentalis (2022)
-
Reprogrammed anti-tumor NK cells
Nature Methods (2021)