Abstract
Expression of Ikaros family transcription factor IKZF3 (Aiolos) increases during murine eosinophil lineage commitment and maturation. Herein, we investigated Aiolos expression and function in mature human and murine eosinophils. Murine eosinophils deficient in Aiolos demonstrated gene expression changes in pathways associated with granulocyte-mediated immunity, chemotaxis, degranulation, ERK/MAPK signaling, and extracellular matrix organization; these genes had ATAC peaks within 1 kB of the TSS that were enriched for Aiolos-binding motifs. Global Aiolos deficiency reduced eosinophil frequency within peripheral tissues during homeostasis; a chimeric mouse model demonstrated dependence on intrinsic Aiolos expression by eosinophils. Aiolos deficiency reduced eosinophil CCR3 surface expression, intracellular ERK1/2 signaling, and CCL11-induced actin polymerization, emphasizing an impaired functional response. Aiolos-deficient eosinophils had reduced tissue accumulation in chemokine-, antigen-, and IL-13–driven inflammatory experimental models, all of which at least partially depend on CCR3 signaling. Human Aiolos expression was associated with active chromatin marks enriched for IKZF3, PU.1, and GATA-1-binding motifs within eosinophil-specific histone ChIP-seq peaks. Furthermore, treating the EOL-1 human eosinophilic cell line with lenalidomide yielded a dose-dependent decrease in Aiolos. These collective data indicate that eosinophil homing during homeostatic and inflammatory allergic states is Aiolos-dependent, identifying Aiolos as a potential therapeutic target for eosinophilic disease.
Similar content being viewed by others
Introduction
Eosinophils are predominantly tissue-dwelling cells that participate in host defense, immunomodulation, and tissue homeostasis.1,2,3 Although commonly assessed in the peripheral blood, the majority of eosinophils leave the circulation and migrate into specific tissues, with the gastrointestinal tract serving as their largest reservoir. Prior to circulation and recruitment to the tissue, eosinophilopoiesis originates from a granulocytes/macrophage progenitor (GMP) precursor, followed by maturation of eosinophil lineage-committed progenitor cells (EoPs). Eosinophil development occurs in the bone marrow via decisive steps in cell fate driven by the action of primary lineage-determining transcription factors (TFs), including GATA-1, GATA-2, PU.1, CEBPα, FOG1, and IRF8.4,5,6,7,8 Subsequently, the EoP lineage commitment is reinforced, either in the bone marrow or tissue, by secondary TFs that orchestrate gene expression, differentiation, and maturation into eosinophils, including the TFs CEBPε and XBP1.9,10,11
More recently, the Ikaros zinc-finger (IKZF) family of TFs was found to have a role in eosinophil lineage commitment and maturation.12 The IKZF TF family includes IKZF1 (Ikaros), IKZF2 (Helios), IKZF3 (Aiolos), IKZF4 (Eos), and IKZF5 (Pegasus), which were first identified for their importance in lymphocyte development and function.13,14 We previously demonstrated that Ikaros, Helios and Aiolos are expressed during eosinophilopoiesis, with Helios and Aiolos being upregulated during eosinophil lineage commitment and maturation.12 Furthermore, chromatin immunoprecipitation coupled with massively parallel sequencing (ChIP-seq) was performed to identify regions of chromatin with active histone modifications. Traditionally, promoters are identified by histone H3 lysine 4 trimethylation (H3K4me3) located proximal (within 1 kB) to the transcription start site (TSS). Enhancers are often identified by open regions, determined using an assay for transposase-accessible chromatin (ATAC)-seq, that are lacking H3K4me3 and are distal (>1 kB) to the TSS.15,16,17,18 Subsequently, enhancers can be classified as poised (lacking enriched acetylation of histone H3 lysine 27 (H3K27ac)) or active (enriched for H3K27ac).19 Notably, Aiolos-binding sites are significantly enriched within the H3K4me3+ promoter region of genes that were expressed during eosinophil maturation, highlighting a potential, novel role for these transcriptional regulators in modulating gene expression during eosinophil development.12 In addition to their role in development, IKZF family members regulate cell migration and cancer metastasis by disrupting cell–cell and cell–epithelial interactions and by decreasing gene expression of the chemokine receptor, integrin, and tight junction genes.20,21 These findings suggest a potentially critical role for IKZF family members in eosinophil functional responses, in addition to those for eosinophil lineage commitment and maturation. However, the involvement of IKZF members in eosinophil migration has yet to be investigated.
Under homeostatic conditions, eosinophil recruitment is regulated by constitutive expression of the G protein-coupled receptor CCR3 on eosinophils and expression of its ligand CCL11 (eotaxin-1) within the gastrointestinal tract.22 Similarly, under inflammatory conditions, an array of chemotactic proteins participate in eosinophil recruitment to the site of inflammation via CCR3.23 Rapid CCR3-mediated eosinophil recruitment and migration to the tissue occurs as a consequence of intracellular activation of the mitogen-activated protein kinases (MAPK) pathway, which has been demonstrated to be involved in multiple aspects of eosinophil biology, including migration, chemotaxis, degranulation, actin polymerization, and autocrine release of multiple inflammatory mediators.24,25,26,27 In addition, numerous clinical and pre-clinical studies have established that CCR3 and its ligands are critical for disease-associated eosinophilia. Considering, (1) the crucial role of CCR3 for homeostatic and inflammatory eosinophil migration, (2) the selective upregulation of Aiolos during eosinophil lineage commitment and development, and (3) the role of Aiolos and IKZF family members in regulating migration in other cell types, we hypothesize that Aiolos not only regulates eosinophil development and maturation but also may regulate downstream functions, such as eosinophil migration potentially through CCR3-mediated receptor signaling.
Herein, we demonstrate that murine eosinophils deficient in Aiolos have marked changes in gene expression, particularly in genes involved in granulocyte-mediated immunity, chemotaxis, intracellular signaling and extracellular matrix organization, and that these genes have ATAC-seq peaks within their promoter regions that were enriched for Aiolos-binding motifs. Aiolos deficiency reduced eosinophil frequency within peripheral tissues, and bone marrow chimera experiments demonstrated dependence on intrinsic Aiolos expression by eosinophils. Furthermore, Aiolos deficiency reduced eosinophil CCR3 protein expression, CCL11-induced intracellular ERK1/2 signaling, and actin polymerization, emphasizing an impaired functional response in Aiolos-deficient eosinophils in response to CCL11/CCR3-mediated signaling. Aiolos-deficient eosinophils had impaired recruitment to tissue in multiple CCR3-dependent experimental models.28,29 Finally, human Aiolos expression was associated with active chromatin marks enriched for IKZF3, PU.1, and GATA-1-binding motifs within eosinophil-specific histone mark ChIP-seq peaks, which were absent in neutrophils. Furthermore, treatment of an immortalized human eosinophilic cell line (EOL-1) with lenalidomide resulted in a dose-dependent decrease in Aiolos levels. These collective data indicate that eosinophil homing during homeostatic and inflammatory allergic states is Aiolos-dependent and identify Aiolos as a potential therapeutic target for eosinophilic disease.
Results
Aiolos deficiency impairs murine eosinophil functional responses
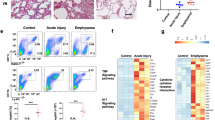
We first examined the expression of Ikaros family members in purified murine tissue-residing eosinophils under homeostatic conditions. Aiolos mRNA was more abundant than Ikaros, Helios, or Gata1 mRNA in murine eosinophils sorted from the bone marrow (BM), and the expression level of all three Ikaros family members declined in the tissue-resident eosinophils (Fig. 1a). Therefore, to determine the functional effect of Aiolos deficiency on eosinophil gene expression, murine bone marrow-derived eosinophils (bmEos) from WT and Aiolos-deficient (Aiolos KO) mice were generated. CCR3 was significantly reduced on the surface of bmEos with a concomitant increase in Siglec-F (Supplementary Fig. 1A). An increase in CD69 and CD11b was also detected on the surface of Aiolos KO bmEos (Supplementary Fig. 1B). No difference was observed in the morphology (Fig. 1b), maturation (Fig. 1c), or viability (Supplementary Fig. 1C) at day 14 of culture between genotypes. Expression of Aiolos mRNA was significantly reduced in Aiolos KO mice, with Aiolos protein detected in WT but not Aiolos KO bmEos (Fig. 1d).
a Expression levels (mean RPKM) of Ikaros family members and Gata1 in murine eosinophils sorted from the bone marrow (BM), intestine (GI), and lung are shown, with CD4+ memory T cells. b Cytocentrifuge preparations (X40) showing morphology of mature bmEos from WT (top) and Aiolos KO (bottom). c Maturation rate of bmEos in culture between genotypes, with representative flow plot showing the proportion of double-positive (Siglec-F+[SigF+]/CCR3+) eosinophils present in culture after 14 days. d Expression levels (FPKM) of Ikaros family mRNA in bmEos generated from WT and Aiolos KO mice (left) n = 5/group, with western blot (right) probed for Aiolos and β-actin, n = 3/group, three independent experiments. e Heatmap showing differentially regulated genes between WT and Aiolos KO mice; downregulated genes are shown in blue, and upregulated genes are shown in red, n = 5/group, three independent experiments. f Gene ontology (GO) analysis highlighting differentially regulated gene pathways between WT and Aiolos KO mice. g Violin plot showing transcription factor (TF) enrichment scores (motif score is the log-odds score of the motif matrix) for Aiolos (IKZF3) motifs in ATAC peaks, as identified using HOMER, within 1 kB of the transcription start site (TSS) of the 371 differentially expressed genes between WT and Aiolos KO mice (i.e., differentially regulated in Aiolos KO) compared to all expressed genes.
Following confirmation of functional Aiolos loss, bmEos were subjected to RNA-seq. A total of 371 genes were differentially regulated (P <0.05, fold change >2) between WT and Aiolos KO mice (Fig. 1e). Differentially regulated genes were enriched for gene pathways associated with granulocyte-mediated immunity, chemotaxis, degranulation, ERK/MAPK signaling, and extracellular matrix organization (Fig. 1f). To determine the activity of Aiolos in the regulation of IKZF3 target genes, we first identified active chromatin regions within 1 kB of the TSS. Active chromatin marks were present in a comparable proportion of dysregulated genes between genotypes (WT: 87.27% ± 3.4, Aiolos KO: 87.16% ± 4.7). Furthermore, ATAC peaks within 1 kB of the TSS of these 371 genes (white violin) were significantly enriched for Aiolos-binding motifs compared to ATAC peaks of all other expressed genes (gray violin) (P <0.0001; Fig. 1g).
Baseline tissue eosinophil accumulation is partially Aiolos-dependent
To determine the functional effect of Aiolos deficiency, we measured the eosinophil frequency in different tissues at homeostasis. Aiolos KO mice have a similar percentage of BM eosinophils to that of WT mice, n = 16–18/group. Peripheral blood eosinophilia (P = 0.0019) was noted in the Aiolos KO mice at homeostasis, n = 16–18/group, with a concomitant ~30–50% decrease in the frequency of eosinophils residing in the intestinal lamina propria (P = 0.0224, n = 10/group) and lungs (P = 0.0167, n = 6/group) (Fig. 2a). To determine whether the reduced frequency of eosinophils in the peripheral tissues of the global Aiolos KO mice was intrinsic to the hematopoietic compartment, we reconstituted sub-lethally irradiated CD45.1+ mice with CD45.2+ WT or Aiolos KO BM cells (Fig. 2b). There was no difference between WT and Aiolos KO eosinophil production in chimera BM (Fig. 2b, left). In contrast, repopulation of the intestine was reduced by ~50% (P = 0.0217, n = 4/group) in Aiolos KO eosinophils compared to WT eosinophils (Fig. 2b, right). Collectively, these data suggest that eosinophil homing to tissues during homeostasis is at least partially dependent upon Aiolos expression by eosinophils.
a Representative flow plots with frequency (mean ± SEM) of eosinophils in the bone marrow (n = 16–18 mice/group, >5 independent experiments), blood (n = 16–18 mice/group, >5 independent experiments), small intestine (n = 10 mice/group, three independent experiments), and lung (n = 6 mice/group, two independent experiments) of wild-type (WT) or Aiolos-deficient (Aiolos KO) mice are shown. b Experimental schema (top) for adoptive transfer of CD24.2 WT or Aiolos KO whole bone marrow cells into irradiated CD45.1-recipient mice. Representative contour plots showing mature eosinophils in the bone marrow (left) and small intestine (right) of CD45.1+ WT mice 4 weeks after transplantation with WT (CD45.2+) or Aiolos KO (CD45.2+) bone marrow are shown. Percentage of live, Siglec-F+[SigF+]CCR3+, and CD45.2+ cells from the donor bone marrow is shown in the upper right. Bar charts showing frequency (mean ± SEM) of donor eosinophils in the bone marrow and small intestine of the recipient mice (n = 4 mice/group, three independent experiments) are shown. ns not significant, SigF+ Siglec-F+.
Aiolos deficiency reduces CCR3 protein expression
To investigate the mechanism underlying the homeostatic migratory defect observed in Aiolos KO eosinophils, we initially measured CCR3 surface expression on murine peripheral blood eosinophils. CCR3 protein expression was reduced on the surface of peripheral blood Aiolos KO eosinophils compared to WT eosinophils (Fig. 3a, top left). We also noted a reduction of CCR3 surface expression on the gastrointestinal tract and lung resident eosinophils in Aiolos KO compared to WT mice (Fig. 3a; small intestine, top right; lung, bottom left). CCR3 surface expression was not different between genotypes in the BM (Fig. 3a, bottom right). A concurrent increase in Siglec-F expression was observed in the BM, peripheral blood and gastrointestinal eosinophils, unlike in the lungs (Supplementary Fig. 2A); however, no biologically relevant difference in the viability of primary eosinophils isolated from the BM and small intestine was observed between genotypes, n = 3/group (Supplementary Fig. 2B). No significant difference was observed in CCR3 mRNA levels, n = 5/group (Fig. 3b).
a CCR3 surface expression, with representative histograms, (geometric mean of fluorescence intensity, mean ± SEM) on wild-type (WT, black circles) and Aiolos-deficient (Aiolos KO, white squares) eosinophils in the bone marrow (n = 16–18 mice/group, >5 independent experiments), blood (n = 16–18 mice/group, >5 independent experiments), small intestine (n = 10 mice/group, three independent experiments), and lung (n = 6 mice/group, two independent experiments) during homeostasis is shown. b CCR3 mRNA gene expression between genotypes n = 5/group, three independent experiments. c Chromatin landscape schema for open (ATAC marked) and active (H3K27ac marked) regions within 20 kB of the murine Ccr3 gene. The red box (middle) identifies an eosinophil promoter located within 1 kB of the transcriptional start site (TSS) of CCR3. The orange boxes (left and right) identify active enhancers identified within 20 kB of CCR3. The colored boxes below the alignment tracks identify shared regulatory elements between genotypes for each chromatin modification; ATAC (black), H3K27ac (blue). The fold change (mean ± SEM) in (d) phosphorylated ERK1/2 (pERK1/2) and (e) actin polymerization in WT (black circles) or Aiolos KO (white squares) eosinophils stimulated with three doses of CCL11 (2, 20, and 200 ng/mL) (n = 3 mice/group, three independent experiments).
We further characterized the surface marker phenotype of primary murine WT and Aiolos KO eosinophil from BM and peripheral blood. CD125 (IL-5Rα) was significantly increased (P = 0.0108) on eosinophils from the BM of Aiolos KO mice, with a nonsignificant trend observed in the peripheral blood (Supplementary Fig. 2C). Integrin subunit beta (ITGβ) 7 was found to be significantly reduced on the surface of peripheral blood eosinophils (P < 0.0001), with the opposite trend (nonsignificant) observed in CD18, CD69, CD11b, and ITGβ2 expression (Supplementary Fig. 2C). Collectively, these data demonstrate the global changes in receptor expression associated with Aiolos deficiency.
MAnorm analysis, a normalization method30 that enables quantitative comparison of ChIP-seq datasets by identifying shared or common peaks based on differential binding, of n = 3–4/group independent, pooled ATAC-seq and ChIP-seq datasets from murine bmEos (Supplementary Fig. 1D) revealed a similar chromatin landscape within 20 kB of the Ccr3 gene body (Fig. 3c) between genotypes. Interestingly, of the three epigenetic peaks shared between genotypes, only the proximal active enhancers (orange boxes) were enriched for Aiolos-binding motifs. Aiolos-binding motifs were not enriched within the shared promoter (red box) identified at the Ccr3 TSS.
CCR3 responses are Aiolos-dependent
CCR3 activation and internalization trigger a downstream cascade of intracellular changes, such as the phosphorylation of signaling molecules and actin polymerization.27 Both CCL11 ligand-induced activation of ERK1/2 and actin polymerization were reduced in Aiolos KO eosinophils compared to WT eosinophils, n = 3/group (Fig. 3d, e). In an attempt to further interrogate the ERK1/2 signaling defect observed in Aiolos KO mice, we questioned whether other activators of MAPK signaling showed similar changes in Aiolos-deficient eosinophils or whether this finding was specific to cytokine stimulation through CCR3. Eosinophils were stimulated with 20 ng/mL CCL11, IL-33, IL-4, and TNFα over an extended time course (0–60 min). Strong phosphorylation of ERK1/2 was detected following stimulation and peaked between 5 and 15 min post-stimulation (Supplementary Fig. 3A). Impaired ERK1/2 phosphorylation was only observed in Aiolos KO mice when eosinophils were stimulated with CCL11 (P = 0.0107) (Supplementary Fig. 3A).
Upon ligation, CCR3 is rapidly internalized, and surface expression is replenished via de novo protein synthesis and recycling of the internalized receptor back to the surface.27 As CCR3 internalization is critical for optimal eosinophil chemotaxis in response to ligands, we hypothesized that CCR3 internalization and surface replenishment may be impaired in Aiolos KO eosinophils following mobilization into the bloodstream or recruitment into tissues. Within 15 min of CCL11 exposure, CCR3 surface expression was reduced by 50%, and homeostatic surface expression was restored within 18 h of ligand induction in both genotypes, n = 3/group. Notably, there was no difference between Aiolos KO and WT eosinophils in ligand-induced internalization or surface CCR3 replenishment at any CCL11 dose tested (Supplementary Fig. 3B), suggesting that reduced CCR3 recycling is not responsible for the impaired chemotactic response observed in Aiolos KO eosinophils.
CCL11-mediated eosinophil accumulation in vitro and in vivo are Aiolos-dependent
Because the chemokine receptor CCR3 and its ligands are critical for eosinophil homing, we investigated the migratory capacity of Aiolos KO and WT eosinophils to the CCR3 ligand CCL11. Migration in vitro toward CCL11 was reduced in Aiolos KO eosinophils compared to WT eosinophils, n = 3/group (Fig. 4a). Furthermore, eosinophil accumulation into the peritoneal cavity 3 h following intraperitoneal (i.p.) injection of CCL11 (1 μg) was reduced by >70% (P = 0.0073) in the Aiolos KO compared to WT mice (Fig. 4b, left). No significant difference was observed between the proportion of eosinophils in the peripheral blood between genotypes (Fig. 4b, right), n = 6/group. Collectively, these data highlight an impaired migratory response to the chemotactic factor CCL11 in eosinophils with Aiolos deficiency.
a Total wild-type (WT, black circles) and Aiolos KO (white squares) eosinophils (mean ± SEM) that migrated toward CCL11 in vitro at the indicated doses of CCL11 is shown (n = 3 mice/group, three independent experiments). b Total (mean ± SEM) WT (black circles) and Aiolos KO (white squares) peritoneal (left) and peripheral blood (right) eosinophils 3 h after intraperitoneal (i.p.) administration of CCL11 (1 μg) is shown (n = 6 mice/group, three independent experiments). c Experimental schema (top) for adoptive transfer of CD45.2 WT or Aiolos KO bmEos into CD45.1 WT mice, following four intranasal (i.n.) OVA challenge. Ratio (left) (mean ± SEM) and absolute count (right) of adoptively transferred (CD45.2+) WT or Aiolos KO eosinophils to native (CD45.1+) WT eosinophils in the bronchoalveolar lavage fluid (BALF) 24 h after final allergen challenge (n = 6–7 mice/group, three independent experiments). d Experimental schema (top) for doxycycline-induced eosinophil accumulation in CC10-iIL-13–WT and CC10-iIL-13–Aiolos KO mice. Percentage (mean ± SEM) of BALF cells that were eosinophils in WT (black circles) and Aiolos KO (white squares) bitransgenic mice (CC10-iIL-13) that were fed doxycycline-impregnated food for 2 weeks (n = 9–11 mice/group, four independent experiments).
Allergen-induced pulmonary eosinophil accumulation in vivo is Aiolos-dependent
As the global Aiolos KO mice may have lymphocyte defects that may affect allergen sensitization,31 we adoptively transferred WT and Aiolos KO CD45.2+ bmEos into ovalbumin (OVA)-sensitized CD45.1+ WT mice after allergen challenge and measured the accumulation of CD45.2+ eosinophils into the BALF after 24 h (Fig. 4c, top). Notably, accumulation of Aiolos KO eosinophils in the airway in response to allergen challenge was reduced by >50% (P = 0.0009) compared to that of WT eosinophils (Fig. 4c, bottom left), with a significant reduction (P = 0.0301) observed in the absolute number of recruited CD45.2 eosinophils to the lungs (Fig. 4c, bottom right).
IL-13-induced pulmonary eosinophil accumulation in vivo is Aiolos-dependent
In a separate lung eosinophilia model, we tested the response of Aiolos-deficient eosinophils to pulmonary overexpression of IL-13 by generating inducible IL-13 transgenic mice deficient in Aiolos (Fig. 4d, top). Eosinophil accumulation in the airway of mice following 2 weeks of pulmonary-restricted expression of an inducible IL-13 transgene was reduced by >50% (P = 0.0098) in Aiolos KO (CC10-iIL-13–Aiolos KO) mice compared to WT (CC10-iIL-13–WT) mice (Fig. 4d, bottom), n = 9–11/group.
Human granulocyte-specific expression of Aiolos is restricted to eosinophils
As data presented in this manuscript, and our previous work investigating the role of Aiolos in eosinophil maturation,12 were performed exclusively in murine eosinophils or using murine inflammatory models, we wanted to validate the importance of Aiolos in human eosinophils. Protein lysates and mRNA from human granulocytes isolated from the peripheral blood of non-atopic patients (Fig. 5a) were subjected to analysis. Protein expression of Aiolos was detectable in human eosinophils but absent in neutrophils (Fig. 5b). Furthermore, gene expression analysis revealed that human eosinophils express Ikaros, Helios, and Aiolos mRNA, whereas neutrophils express Ikaros, but not Helios or Aiolos mRNA (Fig. 5c and Supplementary Fig. 4A, B). Despite clear expression of Aiolos by human eosinophils, the absolute expression levels of these factors relative to lymphocytes were low (Fig. 5d).
a Representative images (×40) of human peripheral blood-derived neutrophils (left) and eosinophils (right) isolated for downstream analysis. b Human eosinophils and neutrophils isolated from peripheral blood of healthy control subjects probed for Aiolos (IKZF3) and β-actin, n = 2, two independent experiments. c Relative human neutrophil and eosinophil gene expression, normalized to GAPDH, with chromatin landscape for the H3K27ac and H3K4me3 histone marks assessed by ChIP-seq proximal to IKZF3 gene—in human eosinophils and neutrophils. The red box identifies eosinophil-specific peaks located within 1 kB of the TSS. A scatter plot showing normalized gene expression relative to GAPDH (mean ± SEM) of IKZF3 between human eosinophils and neutrophils isolated from the peripheral blood of individual human donors is shown at the left. d Expression levels (mean RPKM) of Ikaros family members and Gata1 in human eosinophils, CD4+ T cells, and Jurkat cell line. e TF enrichment scores for PU.1, IKZF1/2/3, and GATA-1 motifs within the IKZF1, IKZF2, and IKZF3 gene promoter region (±1 kB) ChIP-seq peaks (mean ± SEM, for all group). ac acetylation, H histone, K lysine, me3 trimethylation, TSS transcription start site.
Functional genomics analysis for histone modifications H3K4me3 and H3K27ac on three independent human donors (Supplementary Fig. 4C) revealed that the epigenetic regulation of the chromatin landscape mirrored the gene expression profile at Ikaros family gene loci. MAnorm analysis revealed eosinophil-specific histone peaks (red box) within 1 kB of the TSS of Aiolos (Fig. 5c) and Helios (Supplementary Fig. 4A); these peaks were absent in human neutrophils. Ikaros, which is expressed by both granulocyte populations, possessed a common granulocyte histone peak (black box) within 1 kB of the TSS (Supplementary Fig. 4B). TF-binding motif analysis (HOMER) revealed putative PU.1, IKZF3, and GATA-1-binding sites within the eosinophil-specific histone peaks of Helios and Aiolos, whereas Ikaros lacked a strong predicted GATA-1-binding site (Fig. 5e). Furthermore, an assessment of the chromatin landscape surrounding the human CCR3 gene revealed an eosinophil-specific promoter region (red box) and two eosinophil-specific enhancers (orange boxes) that were not present in neutrophils (Supplementary Fig. 4D). The promoter region of the CCR1 gene located ~33 kB from the TSS of CCR3 possessed a shared granulocytes promoter (black box) for reference. Interestingly, of the three eosinophil-specific epigenetic peaks, only the proximal enhancers (orange boxes) were enriched for Aiolos-binding motifs (Supplementary Fig. 4E). Aiolos-binding motifs were not enriched within the CCR3 promoter (red box). These data support a specific role for Aiolos in human eosinophils, which contrasts with the traditional view of these TFs as lymphocyte restricted. Lastly, treatment of a human eosinophilic cell line (EOL-1) with increasing doses of lenalidomide resulted in a dose-dependent decrease in Aiolos expression (Supplementary Fig. 4F), suggesting that eosinophil Aiolos levels may be targeted for therapeutic treatment of allergic disease.
Discussion
Herein, we demonstrated a novel role for the IKZF family TF Aiolos in regulating eosinophil migration. Although Aiolos may impact multiple aspects of eosinophil migratory behavior, we demonstrated that Aiolos specifically contributed to CCL11/CCR3-mediated migration of eosinophils under homeostatic and inflammatory conditions. We previously reported expression of the Ikaros family of TFs in murine eosinophils isolated from the bone marrow, with significant enrichment of predicted Aiolos- and Helios-binding sites within active chromatin of eosinophils and EoPs, supporting a critical role during eosinophilopoiesis.12 Data presented herein further substantiate the differential expression of IKZF family isoforms between human granulocyte populations, with eosinophils expressing Ikaros, Helios, and Aiolos and neutrophils expressing only Ikaros. Interestingly, the decrease observed in Aiolos mRNA observed in tissue-resident murine eosinophils, compared with the highest expression level in BM, further highlights the potential role of Aiolos in migration to the peripheral tissues; once the eosinophils leave the BM, they have decreased Aiolos expression. The mRNA levels detected in human peripheral blood eosinophils were similar to those in tissue-resident murine populations, further suggesting that this may be the case. Taken together, these data suggest that the upregulation of Aiolos during development is an important step for human and murine eosinophil cell identity in granulocyte differentiation and function.
To determine the functional role of Aiolos in eosinophil migration to the tissue, we examined Aiolos-deficient mice and demonstrated that the frequency of tissue-resident eosinophils were reduced by Aiolos deficiency by a mechanism dependent upon Aiolos expression in eosinophils. We demonstrated impaired recruitment of Aiolos-deficient eosinophils in response to CCL11, both in vitro and in vivo. As in vivo chemotaxis toward i.p. CCL11 was only performed at one time point (3 h), we used two additional, distinct, chronic in vivo inflammatory models, both of which are at least partially dependent on CCR3 signaling,28,29 to further interrogate the migratory defect observed in Aiolos KO mice. Recruitment of eosinophils to the tissue was significantly reduced in Aiolos KO mice in both a model of adoptive transfer of Aiolos-deficient eosinophils into OVA-challenged WT mice and a model of restricted expression via an inducible IL-13 transgene in the lung (CC10-iIL-13).
Mechanistically, Aiolos-deficient eosinophils had impaired ERK1/2 signaling, a key event involved in multiple aspects of eosinophil chemotaxis and migration. Enrichment for gene pathways related to ERK1/2 and MAPK signaling reinforce our findings showing defective ERK1/2 phosphorylation in Aiolos KO mice. Impaired CCL11-mediated ERK1/2 phosphorylation was observed in a dose-dependent manner; furthermore, this defect was only observed in response to cytokine stimulation with CCL11 and was not observed with IL-4, IL-33, nor TNFα (20 ng/mL) stimulation. Interestingly, studies looking at a loss of IKZF family members in other cell types have similarly identified defective ERK signaling as one consequence for the migratory defect observed in these cells.32 Furthermore, lenalidomide, which is an immunomodulatory agent that promotes the ubiquitination and degradation of specific substrates of an E3 ubiquitin ligase and results in rapid and selective degradation of Ikaros (IKZF1) and Aiolos (IKZF3) in lymphocytes,33 has similarly been shown to reduce phosphorylated ERK1/2.34 We demonstrate here that lenalidomide decreased Aiolos protein expression in the EOL-1 eosinophilic cell line. Thus, Aiolos appears an attractive target for therapeutic strategies due to its selectivity for migration to the tissues.
The loss of Aiolos impacted eosinophil gene expression involved in cellular pathways related to cell migration and intracellular signaling. Our finding parallels those by others demonstrating that Aiolos mediates its effect on chemotaxis by altering gene expression of the chemokine receptor, integrin, and tight junction genes.20,21,35 Indeed, the observed enrichment for IKZF3-binding motifs within the promoter region of the 371 genes differentially regulated between WT and Aiolos KO bmEos further highlights that this subset of genes may crucially depend upon Aiolos for their expression in eosinophils. Interestingly, Ccr3 mRNA was not significantly reduced in Aiolos KO mice, and Aiolos-binding motifs were enriched in the active enhancers located outside the gene body, compared to the within the promoter region of the Ccr3 gene. These data imply that Aiolos is responsible for post-translational or epigenetic modification of Ccr3 that affects its functions, as demonstrated within this manuscript, rather than the expression of the gene itself. Parallel findings in the epigenetic landscape of the human CCR3 with the eosinophil-specific enhancers, not the eosinophil-specific promoter, being enriched for Aiolos-binding motifs suggest that functional regulation may be similar in humans. Future studies will attempt to determine functional activity at these and other candidate Aiolos (IKZF3)-enriched active enhancers, initially by using massively parallel reporter assays in murine bmEos.
One caveat to this study is that all murine NGS analysis and functional experiments were performed in culture-derived bmEos and not primary eosinophils. Generation of culture-derived bmEos allows for practical utility, enabling the generation of a large yield of cells (~20–40 million/mouse) that can be used for downstream analysis. As bmEos are generated in the presence of super-physiological levels of mouse IL-5, they may not represent naïve, homeostatic eosinophils but rather active, tissue-recruited populations. As similar trends are observed in the CCR3, Siglec-F, CD18, CD11b, and ITGβ7 surface expression profile of bmEos and in that of primary murine eosinophils found in the peripheral blood, we are confident that these represent an appropriate eosinophil population to native eosinophils.
Taken together, our data support a critical role for Aiolos in the regulation of CCR3-mediated eosinophil migratory responses during homeostasis and inflammatory processes. Future work to interrogate the intracellular signaling defect observed in murine Aiolos KO eosinophils and the impact of Aiolos (IKZF3) expression in allergic disease severity may enable the development of pharmacologic treatment approaches to reduce eosinophil Aiolos levels and provide therapeutic benefit to patients with eosinophilic diseases.
Methods
Mice
Age-matched male and female mice (5–10 weeks, C57BL/6 background) were used for all experiments. Wild-type (WT) and WT/CD45.1 mice were purchased from Jackson Laboratories. Aiolos-deficient (Aiolos KO36) mice were generously provided by Dr. Katia Georgopoulos (Harvard University, MA). Bitransgenic (CC10-iIL-13) mice were generated as previously described.37 CC10-iIL-13/WT or CC10-iIL-13/Aiolos KO were generated by breeding CC10-iIL-13 with Aiolos KO mice. All mice were housed under specific pathogen-free conditions and handled under approved protocols (IACU2018-0028) of the Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital Medical Center (CCHMC).
Isolation of human peripheral blood granulocytes and mononuclear cells (PBMCs)
Heparinized venous blood was obtained from both atopic and non-atopic volunteers (>6 years of age) in accordance with a protocol approved by the Institutional Review Board for Human Subjects at CCHMC (IRB 2008-0090). Granulocytes were isolated using dextran sedimentation followed by density gradient separation as described.38 Following isolation of the granulocyte layer, purified eosinophils were collected by negative selection using the eosinophil-isolation kit (#130-042-901, Mitenyi Biotec). Eosinophil and neutrophil purity was assessed by HEMA-3 staining (23-123869, Fisher Scientific) followed by microscopic analysis, and the suspensions were routinely 95–98% pure for the respective granulocyte.
Quantitative PCR and mRNA sequencing
For human gene expression studies, the total RNA was isolated from purified cells using TRIzol™ (Invitrogen) followed by purification with the Direct-zol™ RNA MiniPrep (Zymo Research). Reverse transcriptase PCR was completed using the SuperScript™ VILO™ cDNA Synthesis Kit. Quantitative real-time PCR was performed to determine gene expression using TaqMan® Gene Expression Assays. A list of the probes used for these studies is provided in the Supplementary Material (Supplementary Table 1). For murine studies, the total RNA was isolated by double chloroform extraction and purified as above, followed by reverse transcription with the iScript DNA synthesis kit (BioRad). Quantitative real-time PCR was performed with Sybr Green MasterMix (Applied Biosystem) in an ABI Prism 7900 detection system. For mRNA sequencing, RNA quality was assessed via Agilent RNA NanoCHIP, and only samples with an RNA quality number >8 were used. Libraries were constructed with the TruSeq Stranded mRNA kit and sequenced on the NovaSeq 6000 (S1 Flow Cell), paired-end 100 (PE100) by the CCHMC DNA Sequencing and Genotyping Core, Cincinnati, Ohio, targeting ~20 million reads per sample.
Functional genomics assays
A total of 6.5 × 104 viable eosinophils were pelleted at 500×g for 5 min at 4 °C prior to resuspension in 50 μL of ATAC Resuspension Buffer (ATAC-RSB; 10 mM Tris-HCl, 10 mM NaCl, and 3 mM MgCl2 prepared to a total volume of 50 mL in double-distilled water (ddH2O)) with added 0.1% NP-40, 0.1% Tween-20 and 0.01% Digitonin and were processed for assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq), as previously described.39 For chromatin immunoprecipitation with massively parallel DNA sequencing (ChIP-seq), a total of 1–3 × 107 viable eosinophils and/or neutrophils were pelleted in separate tubes, and chromatin was cross-linked by the addition of 1:10 of 10× crosslinking solution (8.8% formaldehyde, 0.1 M NaCl, 1 mM EDTA, 1 mM EGTA, 50 mM HEPES prepared to a total volume of 1 mL in ddH2O) and incubated on ice for 5 min prior to processing as described40 with antibodies for H3K4me3 (17-614, Millipore) and H3K27ac (C15410196, Diagenode).
Functional genomics data analysis
RNA sequencing was performed using the command-line, data-processing pipeline NextGenAligner (Mario Pujato (2019), https://github.com/MarioPujato/NextGenAligner). Briefly, the raw sequence read quality was analyzed using FastQC/0.11.7 (Simon Andrews, FastQC—a quality control application for FastQ files (2018), https://github.com/s-andrews/FastQC). Adapter sequences were trimmed, using CutAdapt/1.8.1 and Trim Galore/0.4.2, prior to alignment of sequence reads to the mm10 genome and RefSeq-based transcriptome using STAR/2.5.41 Duplicates were removed using Picard/1.89 (Broad Institute, Picard Toolkit (2019), http://broadinstitute.github.io/picard). FeatureCounts and calculateFPKMs were used to quantify gene transcript fragments per kilobase per million mapped reads (FPKM). Differential gene expression was determined using DESeq2.42 Gene Ontology (GO) analysis for biological processes, pathways, and molecular functions was performed using Enrichr.43 ATAC and ChIP-sequencing analysis were performed as above; however, reads were aligned to the mm10 or hg19 genome and RefSeq-based transcriptome using BOWTIE/2.3.4.1.44 Duplicates were removed using Picard/1.89, and peaks were called using MACS/2.1.045 with a q value of 0.01 to generate a BED file of peak coordinates. Presence of ATAC-seq or ChIP-seq peaks proximal (within 20 kB) to expressed eosinophil genes was determined using BEDtools/2.27.0.46 ATAC-seq and ChIP-seq tracks were visualized using the Integrative Genomics Viewer (IGV) browser. Differential binding analysis of ChIP-seq and ATAC-seq peaks was performed using DiffBind/2.16.0 (DiffBind: differential binding analysis of ChIP-Seq peak data (2011)).47 MAnorm30 analysis was performed using filtering values of M value = 0.58 and P value = 0.05 to define unique and shared peaks. TF-binding motif enrichment analyses of identified regulatory elements were performed using the HOMER software package.48 Briefly, we created a version of HOMER that uses a library of >7000 TF-binding models (in the form of position weight matrices) taken from the CisBP database49 to scan a set of input sequencing for statistical enrichment of each position weight matrix. Calculations were performed using ZOOPS (zero or one occurrence per sequence) scoring coupled with hypergeometric enrichment analysis to determine motif enrichment. Input eosinophil sequences were also assessed for statistical enrichment of motifs for IKZF, GATA-1, and PU.1-binding sites using the findPeaks program and factor mode within HOMER. Motif score is displayed as log-odds score of the motif matrix. Significantly enriched TF-binding site motifs are expressed as log P values.
Murine bone marrow-derived eosinophils (bmEos)
Bone marrow-derived eosinophils (bmEos) were generated from WT and Aiolos KO mice, as previously reported.50
Identification of tissue and airway eosinophils
Intestinal mononuclear cells were isolated as previously reported.51 For interstitial lung eosinophils, lungs were perfused with phosphate-buffered saline (PBS) and finely chopped prior to enzymatic digestion with LiberaseTL (5401020001, Roche). For airway eosinophils, mice were euthanized, the lungs were flushed twice with 1 mL of PBS with 1% bovine serum albumin (BSA) injected through a catheter inserted in the trachea to collect the bronchoalveolar lavage fluid (BALF), and the BALF was centrifuged at 1200 rpm for 5 min.
Bone marrow chimera
Mice were irradiated using a split dose of 700 rads (7.0 Gy) and a subsequent 475 rads (4.75 Gy) 3 h after. Following irradiation, 10 million bone marrow cells, isolated from WT or Aiolos KO mice, were administered into WT CD45.1 mice by tail vein injection. After 4 weeks, tissues were harvested and stained with the following anti-mouse antibodies: Siglec-F-BV421 (BD Biosciences), CCR3-FITC (R&D), CD45.1-Alexa Fluor 647 (Biolegend), CD45.2-PE-Cy7 (Biolegend), and Zombie NIR fixable viability kit (Biolegend).
CCR3 internalization
In a 96-well round-bottom plate, bmEos were plated at 200,000 cells per well and incubated at 37 °C with or without murine (m)CCL11 (250-01, Peprotech). Cells were stained on ice with anti-CCR3 (FAB729F; R&D) for 30 min in the dark. The fluorescence intensity was evaluated by FACS.
CCR3 signal transduction
To assess phosphorylated extracellular signal-regulated kinase (phospho-ERK), 5 × 105 bmEos were incubated for 10 min at 37 °C. Increasing doses of mCCL11 were added to the wells for 30, 120, and 300 s. Paraformaldehyde (PFA, 1% final) was added to each well for 10 min at 37 °C. Cells were resuspended in PBS, and ice-cold methanol (90% final concentration) was slowly added. Cells were permeabilized overnight at −20 °C and then washed and stained with anti-phospho-ERK1/2 Alexa Fluor 647 antibody (1:500, #13148, Cell Signaling Technologies (CST)) for 60 min at room temperature (RT). To assess F-actin, bmEos were plated at 100,000 cells/well and incubated for 10 min at 37 °C. Cells were stimulated with 2, 20, or 200 ng/mL mCCL11 for 10, 20, 30, or 60 s. After treatment, the cells were incubated with PFA and permeabilized with PBS with 1% BSA and 0.05% TritonX-100. Cells were incubated for 20 min at RT in PBS with 1% BSA containing Phalloidin-Alexa Fluor 488 (Molecular Probes) at a 1:1000.
Western blotting
For cytokine stimulation experiments, bmEos were resuspended at 1 × 106/mL in PBS and rested for 90 min at 37 °C prior to incubation with 20 ng/mL of eotaxin-1, IL-4, IL33, or TNFα for 0–60 min. Eosinophil protein lysates were generated as previously50 and were separated on a BOLT 4–12% gel prior to transfer. Membranes were blocked with Odyssey blocking buffer (#927-50000, LiCor) prior to incubation with primary antibodies directed against Aiolos (1:1000, #15103, CST), phospho-ERK1/2 (1:1000, #4370, CST), ERK1/2 (1:1000, #9107, CST), and β-actin (1:1000, #3700, CST). After incubation with primary antibodies, an appropriate species-specific IRDye secondary antibody was selected (1:10,000, LiCor). Membranes were scanned with the LiCor Odyssey scanning system; the adjusted relative density was calculated using Fiji. The densitometry analysis represents three independent experiments (n = 3). EOL-1 nuclear lysates were generated using the NE-PET Nuclear and Cytoplasmic Extraction kit (#78833, ThermoFisher) and processed as above prior to incubation with primary antibodies directed against Aiolos and Lamin B1 (1:1000; ab133741, Abcam).
Eotaxin binding
A million bmEos were pre-treated with mouse CCL24 (Peprotech) at 200 ng/mL for 15 min on ice. Then, 20 ng/mL of biotinylated-mCCL11 (E8403, Sigma) was added for 30 min. Cells were then incubated in the presence of streptavidin-PE (Invitrogen) at 1:1000 and assessed by FACS.
Chemotaxis assays
bmEos chemotaxis was evaluated in vitro using 96-well, 5.0-μm pore size transwell plates, as described previously.52 In vivo, WT, or Aiolos KO mice received 1 μg of mCCL11 (Peprotech) or saline solution (200 μL/mouse) intraperitoneally. After 3 h, mice were euthanized, and peritoneal lavage was performed with 5 mL of PBS/1% BSA. The cell suspension was collected, and the presence of eosinophils was determined by FACS.
Adoptive transfer of eosinophils in ovalbumin model
The adoptive transfer model was modified from that previously described.53 Briefly, WT CD45.1 mice received an intranasal dose of 50 µg of ovalbumin (OVA)/Alum (Sigma/Pierce chemical) on days 0 and 7 prior to challenge and on days 14 and 16. Six hours after the last challenge, 10 million bmEos from WT CD45.2 or Aiolos KO CD45.2 were diluted in 200 µL of saline solution and transferred by tail vein injection into OVA-treated WT CD45.1 mice. Twenty-four hours later, the mice were euthanized, and the BALF was harvested as described above.
Statistical analysis
Statistics were done by using the Student’;s t test or two-way ANOVA with Tukey’s multiple comparisons. P values <0.05 were considered significant.
References
Mesnil, C. et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J. Clin. Investig. 126, 3279–3295 (2016).
Akuthota, P., Wang, H. & Weller, P. F. Eosinophils as antigen-presenting cells in allergic upper airway disease. Curr. Opin. Allergy Clin. Immunol. 10, 14–19 (2010).
Lee, S. D. & Tontonoz, P. Eosinophils in fat: pink is the new brown. Cell 157, 1249–1250 (2014).
Hirasawa, R. et al. Essential and instructive roles of GATA factors in eosinophil development. J. Exp. Med. 195, 1379–1386 (2002).
van Dijk, T. B. et al. The role of transcription factor PU.I in the activity of the intronic enhancer of the eosinophil-derived neurotoxin (RNS2) gene. Blood 91, 2126–2132 (1998).
Nerlov, C., McNagny, K. M., Döderlein, G., Kowenz-Leutz, E. & Graf, T. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes Dev. 12, 2413–2423 (1998).
Querfurth, E. et al. Antagonism between C/EBPβ and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 14, 2515–2525 (2000).
Roure, C. D. et al. Hematopoietic overexpression of FOG1 does not affect B-cells but reduces the number of circulating eosinophils. PLoS ONE 9, e92836 (2014).
Lekstrom-Himes, J. A. The role of C/EBP(epsilon) in the terminal stages of granulocyte differentiation. Stem Cells 19, 125–133 (2001).
Gombart, A. F. et al. Regulation of neutrophil and eosinophil secondary granule gene expression by transcription factors C/EBP epsilon and PU.1. Blood 101, 3265–3273 (2003).
Bettigole, S. E. et al. The transcription factor XBP1 is selectively required for eosinophil differentiation. Nat. Immunol. 16, 829–837 (2015).
Bouffi, C. et al. Transcription factor repertoire of homeostatic eosinophilopoiesis. J. Immunol. 195, 2683–2695 (2015).
Winandy, S., Wu, P. & Georgopoulos, K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83, 289–299 (1995).
Georgopoulos, K. et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79, 143–156 (1994).
Heinz, S., Romanoski, C. E., Benner, C. & Glass, C. K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015).
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Wang, Z. et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897–903 (2008).
Ernst, J. et al. Systematic analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010).
He, L.-C. et al. Ikaros inhibits proliferation and, through upregulation of Slug, increases metastatic ability of ovarian serous adenocarcinoma cells. Oncol. Rep. 28, 1399–1405 (2012).
Li, X. et al. Aiolos promotes anchorage independence by silencing p66Shc transcription in cancer cells. Cancer Cell. 25, 575–589 (2014).
Hogan, S. P. et al. Eosinophils: biological properties and role in health and disease. Clin. Exp. Allergy 38, 709–750 (2008).
Sabroe, I. et al. Differential regulation of eosinophil chemokine signaling via CCR3 and non-CCR3 pathways. J. Immunol. 162, 2946–2955 (1999).
Kampen, G. T. et al. Eotaxin induces degranulation and chemotaxis of eosinophils through the activation of ERK2 and p38 mitogen-activated protein kinases. Blood 95, 1911–1917 (2000).
Fulkerson, P. C., Zhu, H., Williams, D. A., Zimmermann, N. & Rothenberg, M. E. CXCL9 inhibits eosinophil responses by a CCR3- and Rac2-dependent mechanism. Blood 106, 436–443 (2005).
Boehme, S. A. et al. Activation of mitogen-activated protein kinase regulates eotaxin-induced eosinophil migration. J. Immunol. 163, 1611–1618 (1999).
Zimmermann, N. & Rothenberg, M. E. Receptor internalization is required for eotaxin-induced responses in human eosinophils. J. Allergy Clin. Immunol. 111, 97–105 (2003).
Fulkerson, P. C., Fischetti, C. A. & Rothenberg, M. E. Eosinophils and CCR3 regulate interleukin-13 transgene-induced pulmonary remodeling. Am. J. Pathol. 169, 2117–2126 (2006).
Fulkerson, P. C. et al. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc. Natl Acad. Sci. USA 103, 16418–16423 (2006).
Shao, Z., Zhang, Y., Yuan, G.-C., Orkin, S. H. & Waxman, D. J. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 13, R16 (2012).
Quintana, F. J. et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat. Immunol. 13, 770–777 (2012).
Schwickert, T. A. et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 15, 283–293 (2014).
Krönke, J. et al. Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523, 183–188 (2015).
Zhu, Y. X. et al. Identification of lenalidomide resistance pathways in myeloma and targeted resensitization using cereblon replacement, inhibition of STAT3 or targeting of IRF4. Blood Cancer J. 9, 1–12 (2019).
Hung, J.-J., Kao, Y.-S., Huang, C.-H. & Hsu, W.-H. Overexpression of Aiolos promotes epithelial-mesenchymal transition and cancer stem cell-like properties in lung cancer cells. Sci. Rep. 9, 1–10 (2019).
Wang, J.-H. et al. Aiolos regulates B cell activation and maturation to effector state. Immunity 9, 543–553 (1998).
Fulkerson, P. C., Fischetti, C. A., Hassman, L. M., Nikolaidis, N. M. & Rothenberg, M. E. Persistent effects induced by IL-13 in the lung. Am. J. Respir. Cell Mol. Biol. 35, 337–346 (2006).
Nauseef, W. M. Isolation of human neutrophils from venous blood. Methods Mol. Biol. 412, 15–20 (2007).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Yukawa, M. et al. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J. Exp. Med. 217, e20182009 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Gaspar J. M. Improved peak-calling with MACS2. Preprint at https://www.biorxiv.org/content/10.1101/496521v1.full.pdf (2018).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ross-Innes, C. S. et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38, 576–589 (2010).
Lambert, S. A. et al. Similarity regression predicts evolution of transcription factor sequence specificity. Nat. Genet. 51, 981–989 (2019).
Felton, J. M. et al. Mcl-1 protects eosinophils from apoptosis and exacerbates allergic airway inflammation. Thorax 75, 600–605 (2020).
Lee, J.-B. et al. IL-25 and CD4(+) TH2 cells enhance type 2 innate lymphoid cell-derived IL-13 production, which promotes IgE-mediated experimental food allergy. J. Allergy Clin. Immunol. 137, 1216–1225 (2016).
Schollaert, K. L., Stephens, M. R., Gray, J. K. & Fulkerson, P. C. Generation of eosinophils from cryopreserved murine bone marrow cells. PLoS ONE 9, e116141 (2014).
Wen, T., Besse, J. A., Mingler, M. K., Fulkerson, P. C. & Rothenberg, M. E. Eosinophil adoptive transfer system to directly evaluate pulmonary eosinophil trafficking in vivo. Proc. Natl Acad. Sci. USA 110, 6067–6072 (2013).
Acknowledgements
This project was supported in part by NIH R01 AI130033. We wish to thank Shawna Hottinger for her editorial assistance, Julie Caldwell for the maintenance and breeding of Aiolos-deficient (Aiolos KO) mice, Dr. Mario Pujato for the development of bioinformatic pipelines, and Lee Elizabeth Edsall, Michael Kotliar, Sreeja Parameswaran, and Kevin Ernst for bioinformatic assistance and training. RNA sequencing was performed in collaboration with the Cincinnati Children’s DNA Sequencing and Genotyping Core, Cincinnati, Ohio. Aiolos KO mice were generously provided by Dr. Katia Georgopoulos (Harvard University, MA).
Author information
Authors and Affiliations
Contributions
J.M.F., C.B., K.L.S., J.T.S., A.M., S.V., and B.W. performed experiments. J.M.F., C.B., K.L.S., A.M., J.T.S., M.T.W., and P.C.F. analyzed the data. A.Z.M. and J.T.S. recruited patients and provided samples. L.M. maintained murine colonies. J.M.F., C.B., J.T.S., M.E.R., and P.C.F. wrote the manuscript. All authors reviewed, edited, and approved the manuscript prior to submission.
Corresponding author
Ethics declarations
Competing interests
J.M.F., C.B., K.L.S., A.M., S.V., B.W., A.Z.M., L.M., and M.T.W. have no competing interests. A.B. is a co-founder of Datirium, LLC. P.C.F. and J.T.S. have received research funding from Knopp Biosciences. M.E.R. is a consultant for Pulm One, Spoon Guru, ClostraBio, Serpin Pharm, Allakos, Celgene, Astra Zeneca, Arena Pharmaceuticals, GlaxoSmith Kline, Guidepoint, and Suvretta Capital Management; has an equity interest in the first five listed; and has royalties from reslizumab (Teva Pharmaceuticals), PEESSv2 (Mapi Research Trust), and UpToDate. M.E.R. is an inventor of patents owned by Cincinnati Children’s Hospital.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Felton, J.M., Bouffi, C., Schwartz, J.T. et al. Aiolos regulates eosinophil migration into tissues. Mucosal Immunol 14, 1271–1281 (2021). https://doi.org/10.1038/s41385-021-00416-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-021-00416-4
This article is cited by
-
The transcription factor Aiolos restrains the activation of intestinal intraepithelial lymphocytes
Nature Immunology (2024)