Abstract
Background
Colorectal neuroendocrine carcinomas (CRNECs) are highly aggressive tumours with poor prognosis and low incidence. To date, the genomic landscape and molecular pathway alterations have not been elucidated.
Methods
Tissue sections and clinical information of CRNEC (n = 35) and CR neuroendocrine tumours (CRNETs) (n = 25) were collected as an in-house cohort (2010–2020). Comprehensive genomic and expression panels (AmoyDx® Master Panel) were applied to identify the genomic and genetic alterations of CRNEC. Through the depiction of the genomic landscape and transcriptome profile, we compared the difference between CRNEC and CRNET. Reverse transcription-polymerase chain reaction and immunofluorescence staining were performed to confirm the genetic alterations.
Results
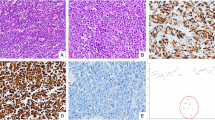
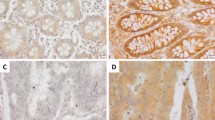
High tumour mutation load was observed in CRNEC compared with CRNET. CRNECs showed a “cold” immune landscape and increased endothelial cell activity compared with NETs. Importantly, PAX5 was aberrantly expressed in CRNEC and predicted a poor prognosis of CRNECs. CCL5, a factor that is considered an immunosuppressive factor in several tumour types, was strongly expressed in CRNEC patients with long-term survival and correlated with high CD8+ T cell infiltration.
Conclusion
Through the depiction of the genomic landscape and transcriptome profile, we demonstrated alterations in molecular pathways and potential targets for immunotherapy in CRNEC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All presented data in this study are available from the corresponding author upon reasonable request.
References
Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020;76:182–8.
Bertani E, Ravizza D, Milione M, Massironi S, Grana CM, Zerini D, et al. Neuroendocrine neoplasms of rectum: a management update. Cancer Treat. Rev. 2018;66:45–55.
Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, et al. Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol. 2017;3:1335–42.
Chen L, Liu M, Zhang Y, Guo Y, Chen M-H, Chen J. Genetic characteristics of colorectal neuroendocrine carcinoma: more similar to colorectal adenocarcinoma. Clin Colorectal Cancer. 2021;20:177–85.e13.
Dizdar L, Werner TA, Drusenheimer JC, Möhlendick B, Raba K, Boeck I, et al. BRAFV600E mutation: a promising target in colorectal neuroendocrine carcinoma. Int J Cancer. 2019;144:1379–90.
Liu L, Zhang R, Deng J, Dai X, Zhu X, Fu Q, et al. Construction of TME and identification of crosstalk between malignant cells and macrophages by SPP1 in hepatocellular carcinoma. Cancer Immunol Immunother. 2021:1–16. https://doi.org/10.1007/s00262-021-02967-8. Online ahead of print
Bao X, Zhang H, Wu W, Cheng S, Dai X, Zhu X, et al. Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy. J Immunother Cancer. 2020;8:e001437.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7.
Aldinucci D, Borghese C, Casagrande N. The CCL5/CCR5 axis in cancer progression. Cancers 2020;12:1765.
Walens A, DiMarco AV, Lupo R, Kroger BR, Damrauer JS, Alvarez JV. CCL5 promotes breast cancer recurrence through macrophage recruitment in residual tumors. Elife 2019;8:e43653.
Huang R, Wang S, Wang N, Zheng Y, Zhou J, Yang B, et al. CCL5 derived from tumor-associated macrophages promotes prostate cancer stem cells and metastasis via activating β-catenin/STAT3 signaling. Cell Death Dis. 2020;11:1–20.
Zazo S, González-Alonso P, Martin-Aparicio E, Chamizo C, Luque M, Sanz-Alvarez M, et al. Autocrine CCL5 effect mediates trastuzumab resistance by ERK pathway activation in HER2-positive breast cancer. Mol Cancer Ther. 2020;19:1696–707.
García-Suárez O, García B, Fernández-Vega I, Astudillo A, Quirós LM. Neuroendocrine tumors show altered expression of chondroitin sulfate, glypican 1, glypican 5, and syndecan 2 depending on their differentiation grade. Front Oncol. 2014;4:15.
Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, et al. Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J. 2012;279:1177–97.
Lindahl U, Kjellén L. Pathophysiology of heparan sulphate: many diseases, few drugs. J Intern Med. 2013;273:555–71.
Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15:1013–31.
Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79.
Souabni A, Jochum W, Busslinger M. Oncogenic role of Pax5 in the T-lymphoid lineage upon ectopic expression from the immunoglobulin heavy-chain locus. Blood 2007;109:281–9.
Robichaud GA, Nardini M, Laflamme M, Cuperlovic-Culf M, Ouellette RJ. Human Pax-5 C-terminal isoforms possess distinct transactivation properties and are differentially modulated in normal and malignant B cells. J Biol Chem. 2004;279:49956–63.
Yan M, Himoudi N, Pule M, Sebire N, Poon E, Blair A, et al. Development of cellular immune responses against PAX5, a novel target for cancer immunotherapy. Cancer Res. 2008;68:8058–65.
Do HTT, Lee CH, Cho J. Chemokines and their receptors: multifaceted roles in cancer progression and potential value as cancer prognostic markers. Cancers. 2020;12:287.
Aldinucci D, Borghese C, Casagrande N. Formation of the immunosuppressive microenvironment of classic Hodgkin lymphoma and therapeutic approaches to counter it. Int J Mol Sci. 2019;20:2416.
Chang L-Y, Lin Y-C, Mahalingam J, Huang C-T, Chen T-W, Kang C-W, et al. Tumor-derived chemokine CCL5 enhances TGF-β–mediated killing of CD8+ T cells in colon cancer by T-regulatory cells. Cancer Res. 2012;72:1092–102.
Jiao X, Velasco-Velázquez MA, Wang M, Li Z, Rui H, Peck AR, et al. CCR5 governs DNA damage repair and breast cancer stem cell expansion. Cancer Res. 2018;78:1657–71.
Wang S-W, Liu S-C, Sun H-L, Huang T-Y, Chan C-H, Yang C-Y, et al. CCL5/CCR5 axis induces vascular endothelial growth factor-mediated tumor angiogenesis in human osteosarcoma microenvironment. Carcinogenesis 2015;36:104–14.
Ban Y, Mai J, Li X, Mitchell-Flack M, Zhang T, Zhang L, et al. Targeting autocrine CCL5–CCR5 axis reprograms immunosuppressive myeloid cells and reinvigorates antitumor immunity. Cancer Res. 2017;77:2857–68.
Yang X, Hou J, Han Z, Wang Y, Hao C, Wei L, et al. One cell, multiple roles: contribution of mesenchymal stem cells to tumor development in tumor microenvironment. Cell Biosci. 2013;3:5.
Casagrande N, Borghese C, Visser L, Mongiat M, Colombatti A, Aldinucci D. CCR5 antagonism by maraviroc inhibits Hodgkin lymphoma microenvironment interactions and xenograft growth. Haematologica 2019;104:564–75.
Cambien B, Richard-Fiardo P, Karimdjee BF, Martini V, Ferrua B, Pitard B, et al. CCL5 neutralization restricts cancer growth and potentiates the targeting of PDGFRβ in colorectal carcinoma. PLoS ONE. 2011;6:e28842.
Üçüncü M, Serilmez M, Sarı M, Bademler S, Karabulut S. The diagnostic significance of PDGF, EphA7, CCR5, and CCL5 levels in colorectal cancer. Biomolecules 2019;9:464.
Halama N, Zoernig I, Berthel A, Kahlert C, Klupp F, Suarez-Carmona M, et al. Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell. 2016;29:587–601.
Dangaj D, Bruand M, Grimm AJ, Ronet C, Barras D, Duttagupta PA, et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell. 2019;35:885–900. e10
de Galarreta MR, Bresnahan E, Molina-Sánchez P, Lindblad KE, Maier B, Sia D, et al. β-Catenin activation promotes immune escape and resistance to anti–PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019;9:1124–41.
Huffman AP, Lin JH, Kim SI, Byrne KT, Vonderheide RH. CCL5 mediates CD40-driven CD4+ T cell tumor infiltration and immunity. JCI Insight. 2020;5:e137263.
Böttcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, et al. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 2018;172:1022–37.e14.
Seo W, Shimizu K, Kojo S, Okeke A, Kohwi-Shigematsu T, Fujii S-I, et al. Runx-mediated regulation of CCL5 via antagonizing two enhancers influences immune cell function and anti-tumor immunity. Nat Commun. 2020;11:1–16.
Mowat C, Mosley SR, Namdar A, Schiller D, Baker K. Anti-tumor immunity in mismatch repair-deficient colorectal cancers requires type I IFN–driven CCL5 and CXCL10. J Exp Med. 2021;218:e20210108.
Funding
This study was supported by grants (nos. 81874143, 31971192) from the National Nature Science Foundation of China and grants (LY21H030005, LQ19H160025, LY19H160040) from the Natural Science Foundation of Zhejiang Province.
Author information
Authors and Affiliations
Contributions
DC: investigation, methodology, writing original draft and writing—review and editing. XB: conceptualisation, methodology, data curation, formal analysis, methodology, visualisation, project administration and writing the original draft. RZ: data curation, formal analysis and writing—review and editing. HZ: data curation, formal analysis and writing—review and editing. ZT: data curation, visualisation and writing—review and editing. LL: data curation, validation and writing—review and editing. YZ: data curation, software and writing—review and editing. MZ: data curation, validation and writing—review and editing. XL: validation and writing—review and editing. SW: validation and writing—review and editing. BL: data curation, validation and writing—review and editing. XC: data curation, validation and writing—review and editing. XC: data curation, validation and writing—review and editing. RJ: formal analysis, visualisation, methodology and writing—review and editing. WF: conceptualisation, supervision, funding acquisition and writing—review and editing. PZ: investigation, visualisation, supervision and writing—review and editing.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the ethics committee of The First Affiliated Hospital, College of Medicine, Zhejiang University. All patients/participants provided informed consent.
Consent for publication
All authors reviewed and approved the manuscript.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, D., Bao, X., Zhang, R. et al. Depiction of the genomic and genetic landscape identifies CCL5 as a protective factor in colorectal neuroendocrine carcinoma. Br J Cancer 125, 994–1002 (2021). https://doi.org/10.1038/s41416-021-01501-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01501-y