Abstract
Purpose
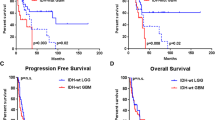
We previously reported that there was a subgroup of IDH-mutated astrocytomas harboring only 19q-loss showing oligodendroglioma-like morphology and significantly longer overall survival (OS) compared with 19q-intact astrocytomas. The aim of this study was to further explore the biological characteristics of this possible subgroup and obtain insight into the mechanism of their relatively benign clinical behavior.
Methods
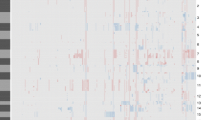
We compared gene expression pattern between five 19q-loss and five 19q-intact IDH-mutated astrocytomas by microarray analysis.
Results
By comparing expression levels of genes of 19q-loss astrocytomas to those of 19q-intact astrocytomas, 102 up-regulated genes and 162 down-regulated genes were extracted. The down-regulated genes clustered heavily to 19q and 4p while the up-regulated genes clustered to 4q. It was noteworthy that fibroblast growth factor 1 associated with stem cell maintenance and multiple genes associated with glioma progression were down-regulated in 19q-loss astrocytomas, and these results were validated with the independent TCGA data set. On t-SNE analysis of the 19q-loss astrocytomas with other IDH-mutant glioma subgroups from the TCGA datasets, the expression pattern of the 19q-loss astrocytomas showed no shift toward oligodendrogliomas with 1p/19q codeletion but rather constituted a subgroup of astrocytoma.
Conclusions
These findings suggested that 19q-loss in astrocytomas is more likely acquired event rather than an early event in oncogenesis like the 1p/19q-codeletion in oligodendrogliomas, and that the biological features of 19q-loss astrocytomas are possibly related to differentially expressed genes associated with stem cell maintenance and glioma progression.


Similar content being viewed by others
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820. https://doi.org/10.1007/s00401-016-1545-1
Otani R, Uzuka T, Higuchi F, Matsuda H, Nomura M, Tanaka S, Mukasa A, Ichimura K, Kim P, Ueki K (2018) IDH-mutated astrocytomas with 19q-loss constitute a subgroup that confers better prognosis. Cancer Sci 109:2327–2335. https://doi.org/10.1111/cas.13635
Kalyani AJ, Mujtaba T, Rao MS (1999) Expression of EGF receptor and FGF receptor isoforms during neuroepithelial stem cell differentiation. J Neurobiol 38:207–224. https://doi.org/10.1002/(sici)1097-4695(19990205)38:2%3C207::aid-neu4%3E3.0.co;2-g
Chiu IM, Touhalisky K, Liu Y, Yates A, Frostholm A (2000) Tumorigenesis in transgenic mice in which the SV40 T antigen is driven by the brain-specific FGF1 promoter. Oncogene 19:6229–6239. https://doi.org/10.1038/sj.onc.1204021
Hsu YC, Kao CY, Chung YF, Lee DC, Liu JW, Chiu IM (2016) Activation of Aurora A kinase through the FGF1/FGFR signaling axis sustains the stem cell characteristics of glioblastoma cells. Exp Cell Res 344:153–166. https://doi.org/10.1016/j.yexcr.2016.04.012
Hide T, Komohara Y, Miyasato Y, Nakamura H, Makino K, Takeya M, Kuratsu JI, Mukasa A, Yano S (2018) Oligodendrocyte progenitor cells and macrophages/microglia produce glioma stem cell niches at the tumor border. EBioMedicine 30:94–104. https://doi.org/10.1016/j.ebiom.2018.02.024
He XX, Du S, Gao SQ, Chen JY, Cao RJ, Xing ZK, Kazim ARS, Yu HL, Zheng QC, Zhu XJ (2018) Humanization of fibroblast growth factor 1 single-chain antibody and validation for its antitumorigenic efficacy in breast cancer and glioma cells. J Cell Mol Med 22:3259–3263. https://doi.org/10.1111/jcmm.13547
Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, Li Z, Liu XB, Li ZQ, Wang ZH, Xue YX (2015) Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget 6:21934–21949. https://doi.org/10.18632/oncotarget.4290
Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, Hovestadt V, Escalante LE, Shaw ML, Rodman C, Gillespie SM, Dionne D, Luo CC, Ravichandran H, Mylvaganam R, Mount C, Onozato ML, Nahed BV, Wakimoto H, Curry WT, Iafrate AJ, Rivera MN, Frosch MP, Golub TR, Brastianos PK, Getz G, Patel AP, Monje M, Cahill DP, Rozenblatt-Rosen O, Louis DN, Bernstein BE, Regev A, Suva ML (2017) Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. https://doi.org/10.1126/science.aai8478
Saurty-Seerunghen MS, Bellenger L, El-Habr EA, Delaunay V, Garnier D, Chneiweiss H, Antoniewski C, Morvan-Dubois G, Junier MP (2019) Capture at the single cell level of metabolic modules distinguishing aggressive and indolent glioblastoma cells. Acta Neuropathol Commun 7:155. https://doi.org/10.1186/s40478-019-0819-y
Huang RQ, Wang SQ, Zhu QB, Guo SC, Shi DL, Chen F, Fang YC, Chen R, Lu YC (2019) Knockdown of PEBP4 inhibits human glioma cell growth and invasive potential via ERK1/2 signaling pathway. Mol Carcinog 58:135–143. https://doi.org/10.1002/mc.22915
Huang RQ, Shi DL, Huang W, Chen F, Lu YC (2016) Increased expression of phosphatidylethanolamine-binding protein 4 (PEBP4) strongly associates with human gliomas grade. J Neurooncol 127:235–242. https://doi.org/10.1007/s11060-015-2040-6
Yang Z, Xie Q, Hu CL, Jiang Q, Shen HF, Schachner M, Zhao WJ (2017) CHL1 is expressed and functions as a malignancy promoter in glioma cells. Front Mol Neurosci 10:324. https://doi.org/10.3389/fnmol.2017.00324
Wu C, Su J, Wang X, Wang J, Xiao K, Li Y, Xiao Q, Ling M, Xiao Y, Qin C, Long W, Zhang F, Pan Y, Xiang F, Liu Q (2019) Overexpression of the phospholipase A2 group V gene in glioma tumors is associated with poor patient prognosis. Cancer Manage Res 11:3139–3152. https://doi.org/10.2147/CMAR.S199207
Kirk IK, Weinhold N, Belling K, Skakkebaek NE, Jensen TS, Leffers H, Juul A, Brunak S (2017) Chromosome-wise protein interaction patterns and their impact on functional implications of large-scale genomic aberrations. Cell Syst 4:357–364. https://doi.org/10.1016/j.cels.2017.01.001
Sarnataro S, Chiariello AM, Esposito A, Prisco A, Nicodemi M (2017) Structure of the human chromosome interaction network. PLoS ONE 12:e0188201. https://doi.org/10.1371/journal.pone.0188201
Acknowledgements
We are grateful to the patients for the donation of their tissue for research and acknowledge the excellent technical assistance of Ms. Kayoko Iwata of the Dokkyo Medical University laboratory.
Funding
This work was supported by a MEXT KAKENHI Grant to R.O (No. JP18K15286).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation was performed by RO, TU, FH, and KU. Data analysis was performed by RO, AM, and KU. The first draft of the manuscript was written by RO and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical approval
All experiments using human samples were approved by the ethics committee at Dokkyo Medical University (No.1431 and 24053).
Consent to participate
Written informed consent was obtained from all individual participants included in the study.
Consent for publication
Patients signed informed consent regarding publishing their data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Otani, R., Mukasa, A., Uzuka, T. et al. Gene expression profiling of 19q-loss astrocytomas suggest a specific pattern associated with the better prognosis. J Neurooncol 154, 221–228 (2021). https://doi.org/10.1007/s11060-021-03816-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-021-03816-5