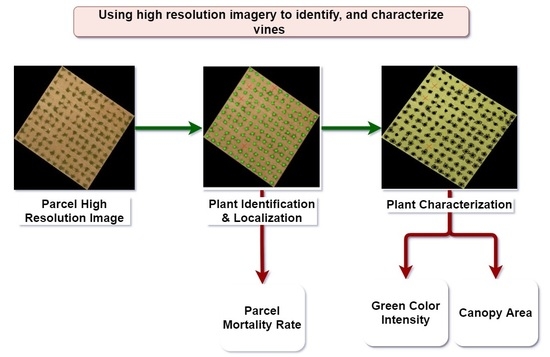
1. Introduction
The rapid evolution of new technologies in precision viticulture allows better vineyard management, monitoring and control of spatio-temporal crop variability; thus helps increasing their oenological potential [
1,
2]. Remote sensing data and image processing techniques are used to fully characterize vineyards starting from automatic parcel delimitation to plant identification.
Missing plant detection has been the subject of many studies. There is a permanent need to identify vine mortality in a vineyard in order to detect the presence of diseases causing damage and, more importantly, as a way of estimating productivity and return on investment (ROI) for each plot. The lower the mortality rate, the higher the ROI. Therefore, mortality rate can help management take better informed decisions for each plot.
Many researchers worked on introducing smart viticulture practices in order to digitize and characterize vineyards. For instance, frequency analysis was used to delimitate vine plots and detect inter-row width and row orientation while providing the possibility of missing vine detection [
3,
4]. Another approach uses dynamic segmentation, Hough space clustering and total least squares techniques to automatically detect vine rows [
5]. In [
6], segmenting the vine rows in virtual shapes allowed the detection of individual plants, while the missing plants are detected by implementing a multi-logistic model. In [
7], the use of morphological operators made dead vine detection possible. In [
8], the authors compared the performance of four classification methods (K-means, artificial neural networks (ANN), random forest (RForest), and spectral indices (SI)) to detect canopy in a vineyard trained on vertical shoot position.
Most of the previous studies concern trellis trained parcels. However, a lot of vine plots adopt the goblet style where vines are planted according to a regular grid with constant inter-row and inter-column spacing. Even though it is an old training style for vineyards, it is still popular in warm and dry regions because it keeps grapes in the shadow, avoiding sunburn that deteriorates grape quality [
9]. Nevertheless, limited research on vine identification and localization were conducted on goblet parcels. A method for localizing missing olive and vine plants in squared-grid patterns from remotely sensed imagery is proposed in [
10] by considering the image as a topological graph of vertices. This method requires the knowledge of the grid orientation angle and the inter-row spacing.
The approach presented in this paper addresses the problem of living and missing vine identification, as well as vine characterization in goblet trained parcels using high resolution aerial images. It is an unsupervised and fully automated approach that requires only the parcel image as input. In the first stage, using the proper image processing techniques, the location of each living and missing vine is determined. In the second stage, a marker-controlled watershed segmentation allows to fully characterize living vines by recognizing their pixels.
Neural networks based methods, more precisely convolutional neural networks (CNN), are used recently and intensively for image processing tasks. Some of these tasks include: image classification to recognize the objects in an image [
11], object detection to recognize and locate the objects in an image by using bounding boxes to describe the target location [
12,
13,
14,
15], semantic segmentation to classify each pixel in the image by linking it to a class label [
16], instance segmentation that combines object detection and semantic segmentation in order to localize the instances of objects while delineating each instance [
17]. All the above-described methods fall in the category of supervised learning. They require learning samples to train the neural network based models. In this study, the CNN-based semantic segmentation is used for comparison purposes.
As outcomes of the proposed approach, a precise mortality rate can be calculated for each parcel. Moreover, living vine characteristics in terms of size, shape, and green color intensity are determined.
3. Results
In order to assess the proposed method, recent ground-truth manual counting is performed on Parcel 59B. Moreover, for all parcels listed in
Table 1, a desktop GIS software is used to manually digitize the vine locations and visually estimate missing vine locations using row and column intersections. For each digitized point, a row and column number is assigned while an attribute of 1 is given to represent a vine and 0 a missing vine. In each case and for each parcel, a matching matrix is computed showing the numbers of truly identified living vines (TLV) and missing vines (TMV), and the numbers of misidentified living vines (FLV) and missing vines (FMV). The accuracy of the proposed method is quantified by calculating the accuracy of missing vines identification (AMV) computed in Equation (
11), the accuracy of living vines identification (ALV) computed in Equation (
12), and the overall accuracy (ACC) computed in Equation (
13).
3.1. Assessment of Proposed Method Compared to Ground-Truth Data
Table 2 shows the comparison between the ground-truth data and the results obtained by applying the proposed method on Parcel 59B. It compares the number of vine rows and the number of living and missing vines in both cases. It also shows the mortality rate in both cases calculated as in Equation (
14).
Figure 16 displays the obtained matching matrix when assessing the proposed method against Parcel 59B’s ground-truth data. Regarding accuracy computation, the accuracy of missing vine identification (AMV) is equal to 74.42%, the accuracy of living vine identification (ALV) is equal to 99.81% and the overall accuracy (ACC) is equal to 99.51%.
3.2. Assessment of Proposed Method Compared to On-Screen Vine Identification
Table 3 shows the values of TLV, TMV, FLV, and FMV. It also shows the accuracy of living vine identification (ALV) (Equation (
12)), the accuracy of missing vine identification (AMV) (Equation (
11)) and the overall accuracy (ACC) (Equation (
13)).
High accuracy values are obtained when comparing the yielded results with ground-truth data and on-screen vine identification. Accuracy values (ACC) exceed 95% for all parcels proving that the proposed method succeeds in identifying missing and living vines. However, the obtained AMV values are lower than the ALV values due to the fact that some small vines, considered dead with on-screen identification, are classified as living vines. Lower accuracy values are obtained when the results are compared with ground-truth data because the parcel may have witnessed many changes since 2017, when the images were taken.
Regarding vine characterization, the pixels of each vine are identified (see
Section 2.2.2). Consequently, further inspection on the size, shape, and green color intensity of each vine can be easily performed. For example, 9.85% of the vines in Parcel 59B have a small size (less than
) and may require special treatment.
Table 4 shows the percentage of vines having a size less than
, between
and
, and greater than
in each parcel. Parcel
60D has the largest percentage of big vines while Parcel
58C has the largest percentage of small vines.
3.3. Comparison with Semantic Segmentation
In order to test the trained DeepLab3vplus model (see
Section 2.3), the image of Parcel 59B (
Figure 1) is presented to the network after resizing it to
pixels where
and
are the closest multiples of 224 to the number of lines and the number of columns of the image, respectively. Even if the size of the test image is different than those of the learning samples, DeepLabv3plus still succeeds in classifying the pixels, as long as the size of the features (vines) are close to the ones learned by the network.
Figure 17 shows the image of Parcel 59B overlayed with the semantic segmentation results using the trained Deeplabv3plus model. It is obvious that the pixels of the image are well classified among four segments: background, soil, plant, and contour.
By setting to one all pixels belonging to the plant segment and to zero all remaining pixels, a binary image is obtained where most of the vines form solitary objects. By applying proper image rotation (
Section 2.2.1.2) and plant identification (
Section 2.2.1.3), the living and missing vines are identified giving an overall accuracy of ACC = 99.5% (Equation (
13)) when these results are assessed with ground-truth data.
Table 5 shows a comparison between the results obtained from the proposed method, those obtained by applying CNN-based Semantic Segmentation followed by plant identification, and those obtained from manual counting on the ground.
4. Discussion
The proposed method succeeded in identifying the living and missing vines of the analyzed parcels with high accuracy (exceeding 95%) giving the possibility to calculate a precise mortality rate. Converting image coordinates to geographical coordinates is possible since each parcel image is a geoTIFF image, which means it is fully georeferenced. A proper intervention on the parcels presenting high mortality rate along with the possibility to locate any missing vine geographically in GIS will increase the parcel’s productivity. Moreover, identifying the pixels of each vine in the context of vine characterization helps detecting any disease that might affect the vines by investigating their size, their shape, and the intensity of their green color. Using CNN-based semantic segmentation instead of K-means clustering yielded quite similar results in terms of vine identification. However, it is a supervised method that requires a large number of learning samples to train the network, whereas the proposed method is unsupervised, requiring only the image as input.
Despite its numerous advantages, this method has some limitations if the vine geometric distribution over the plot grid presents major irregularities. In this case, the sum of rows and the sum of columns signals will fail in detecting the presence of vine rows and vine columns. Additionally, it will be difficult to apply a specific rule for the localization of missing vines. Another limitation may arise from the presence of none-vine plants between the vines that are more likely to belong to the same vine cluster when K-means is used for image segmentation. In this case, one might have recourse to convolutional neural networks based methods that are able to distinguish the vine plants from other plants if the network is well trained. For example, instance segmentation is a potential solution. It produces bounding boxes that surround each instance while recognizing its pixels. Nevertheless, these methods are supervised and need a big number of learning samples that might be unavailable.
5. Conclusions
In this paper, a complete study is presented for vine identification and characterization in goblet-trained vine parcels by analyzing their images. In the first stage, the location of each living and missing plant is depicted. In the second stage, the pixels belonging to each plant are recognized. The results obtained when applying the proposed method on 10 parcels are encouraging and prove its validity. The accuracy of missing and living plants identification exceeds 95% when comparing the obtained results with ground-truth and on-screen vine identification data. Moreover, characterizing each vine helps identifying the leaf size and color for potential disease detection. Additionally, it is an automated method that operates on the image without prior training. Replacing K-means segmentation with CNN-based semantic segmentation yielded good results. However, it is a supervised method that requires network tuning and training.
Parcels delineation methods proposed in literature may be used to automatically crop the parcels images in order to provide a complete and automatic solution for the vineyard digitization and characterization. The success of this method depends on the regular geometric distribution of the vines and on the absence of non-vine plants. Otherwise, supervised methods like instance segmentation might be used for vine identification and characterization with the condition that a learning dataset is available.