Abstract
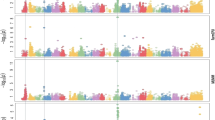
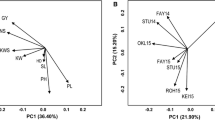
A genome-wide association study (GWAS) for 10 yield and yield component traits was conducted using an association panel comprising 225 diverse spring wheat genotypes. The panel was genotyped using 10,904 SNPs and evaluated for three years (2016–2019), which constituted three environments (E1, E2 and E3). Heritability for different traits ranged from 29.21 to 97.69%. Marker-trait associations (MTAs) were identified for each trait using data from each environment separately and also using BLUP values. Four different models were used, which included three single trait models (CMLM, FarmCPU, SUPER) and one multi-trait model (mvLMM). Hundreds of MTAs were obtained using each model, but after Bonferroni correction, only 6 MTAs for 3 traits were available using CMLM, and 21 MTAs for 4 traits were available using FarmCPU; none of the 525 MTAs obtained using SUPER could qualify after Bonferroni correction. Using BLUP, 20 MTAs were available, five of which also figured among MTAs identified for individual environments. Using mvLMM model, after Bonferroni correction, 38 multi-trait MTAs, for 15 different trait combinations were available. Epistatic interactions involving 28 pairs of MTAs were also available for seven of the 10 traits; no epistatic interactions were available for GNPS, PH, and BYPP. As many as 164 putative candidate genes (CGs) were identified using all the 50 MTAs (CMLM, 3; FarmCPU, 9; mvLMM, 6, epistasis, 21 and BLUP, 11 MTAs), which ranged from 20 (CMLM) to 66 (epistasis) CGs. In-silico expression analysis of CGs was also conducted in different tissues at different developmental stages. The information generated through the present study proved useful for developing a better understanding of the genetics of each of the 10 traits; the study also provided novel markers for marker-assisted selection (MAS) to be utilized for the development of wheat cultivars with improved agronomic traits.








Similar content being viewed by others
Data and materials availability
Not applicable.
References
Allard RW (1999) Principles of Plant Breeding. Wiley and Sons, Inc., New York, p 485
An J, Li Q, Yang J et al (2019) Wheat F-box protein TaFBA1 positively regulates plant drought tolerance but negatively regulates stomatal closure. Front Plant Sci 10:1–20
Astle W, Balding DJ (2009) Population structure and cryptic relatedness in genetic association studies. Stat Sci 24:451–471
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48
Chen K, Li H, Chen Y et al (2015) TaSCL14, a novel wheat (Triticum aestivum L.) GRAS gene, regulates plant growth, photosynthesis, tolerance to photooxidative stress, and senescence. J Genet Genomics 42:21–32
Chen SY, Feng Z, Yi X (2017) A general introduction to adjustment for multiple comparisons. J Thorac Dis 9:1725
Chen Y, Wu H, Yang W et al (2021) Multivariate linear mixed model enhanced the power of identifying genome-wide association to poplar tree heights in a randomized complete block design. G3 Genes Genomes Genet. https://doi.org/10.1093/g3journal/jkaa053
Chu J, Zhao Y, Beier S et al (2020) Suitability of single-nucleotide polymorphism arrays versus genotyping-by-sequencing for genebank genomics in wheat. Front Plant Sci 11:1–12
Cortés J, Mahecha M, Reichstein M, Brenning A (2020) Accounting for multiple testing in the analysis of spatio-temporal environmental data. Environ Ecol Stat 27:293–318
Cui F, Zhao C, Li J et al (2013) Kernel weight per spike: what contributes to it at the individual QTL level? Mol Breed 31:265–278
Cuthbert JL, Somers DJ, Brûlé-Babel AL et al (2008) Molecular mapping of quantitative trait loci for yield and yield components in spring wheat (Triticum aestivum L.). Theor Appl Genet 117:595–608
Daoura BG, Chen L, Du Y, Hu YG (2014) Genetic effects of dwarfing gene Rht-5 on agronomic traits in common wheat (Triticum aestivum L.) and QTL analysis on its linked traits. F Crop Res 156:22–29
Deng X, Wang B, Fisher V et al (2018) Genome-wide association study for multiple phenotype analysis. BMC Proc 12:139–144
Finkelstein RR (2006) Studies of abscisic acid perception finally flower. Plant Cell 18:786–791
Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19:485–494
Furlotte NA, Eskin E (2015) Efficient multiple-trait association and estimation of genetic correlation using the matrix-variate linear mixed model. Genetics 200:59–68
Gahlaut V, Jaiswal V, Singh S et al (2019) Multi-locus genome wide association mapping for yield and its contributing traits in hexaploid wheat under different water regimes. Sci Rep 9:1–15
Gao B, Yang C, Liu J, Zhou X (2021) Accurate genetic and environmental covariance estimation with composite likelihood in genome-wide association studies. PLoS Genet 17:1–25
González JR, Armengol L, Solé X et al (2007) SNPassoc: an R package to perform whole genome association studies. Bioinformatics 23:644–645
Gupta PK, Kulwal PL, Jaiswal V (2014) Association mapping in crop plants: opportunities and challenges. Adv Genet 85:109–147
Gupta PK, Kulwal PL, Jaiswal V (2019) Association mapping in plants in the post-GWAS genomics era. Adv Genet 104:75–154
Gupta PK, Balyan HS, Sharma S, Kumar R (2020) Genetics of yield, abiotic stress tolerance and biofortification in wheat (Triticum aestivum L.). Theor Appl Genet 133:1569–1602
Gyawali A, Shrestha V, Guill KE et al (2019) Single-plant GWAS coupled with bulk segregant analysis allows rapid identification and corroboration of plant-height candidate SNPs. BMC Plant Biol 19:1–15
Henry RJ, Rangan P, Furtado A (2016) Functional cereals for production in new and variable climates. Curr Opin Plant Biol 30:11–18
Hu J, Wang Y, Fang Y et al (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8:1455–1465
IWGSC (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 345:1251788
Jaiswal V, Gahlaut V, Meher PK et al (2016) Genome wide single locus single trait, multi-locus and multi-trait association mapping for some important agronomic traits in common wheat (T. aestivum L.). PLoS ONE 11:1–25
Jamil M, Ali A, Gul A et al (2019) Genome-wide association studies of seven agronomic traits under two sowing conditions in bread wheat. BMC Plant Biol 19:1–18
Juliana P, Singh RP, Poland J et al (2021) Elucidating the genetics of grain yield and stress-resilience in bread wheat using a large-scale genome-wide association mapping study with 55,568 lines. Sci Rep 11:1–15
Kao CH, Zeng ZB, Teasdale RD (1999) Multiple interval mapping for quantitative trait loci. Genetics 152:1203–1216
Kaur S, Zhang X, Mohan A et al (2017) Genome-wide association study reveals novel genes associated with culm cellulose content in bread wheat (Triticum aestivum, L.). Front Plant Sci 8:1–7
Kaya Y, Akcura M (2014) Effects of genotype and environment on grain yield and quality traits in bread wheat (T. aestivum L.). Food Sci Technol 34:386–393
Korte A, Vilhjálmsson BJ, Segura V et al (2012) A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet 44:1066–1071
Kumar N, Kulwal PL, Balyan HS, Gupta PK (2007) QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol Breed 19:163–177
Kumar J, Saripalli G, Gahlaut V et al (2018) Genetics of Fe, Zn, β-carotene, GPC and yield traits in bread wheat (Triticum aestivum L.) using multi-locus and multi-traits GWAS. Euphytica 214:1–17
Kumar A, Sharma S, Chunduri V et al (2020) Genome-wide identification and characterization of heat shock protein family reveals role in development and stress conditions in Triticum aestivum L. Sci Rep 10:1–12
Langer SM, Longin CFH, Würschum T (2014) Flowering time control in European winter wheat. Front Plant Sci 5:1–12
Lee HS, Jung JU, Kang CS et al (2014) Mapping of QTL for yield and its related traits in a doubled haploid population of Korean wheat. Plant Biotechnol Rep 8:443–454
Lemmens M, Scholz U, Berthiller F et al (2005) The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for fusarium head blight resistance in wheat. Mol Plant-Microbe Interact 18:1318–1324
Li Y, Wei K (2020) Comparative functional genomics analysis of cytochrome P450 gene superfamily in wheat and maize. BMC Plant Biol 20:1–22
Lipka AE, Tian F, Wang Q et al (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Liu G, Jia L, Lu L et al (2014) Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor Appl Genet 127:2415–2432
Liu X, Huang M, Fan B et al (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLOS Genet 12:1–24
Muhammad A, Hu W, Li Z et al (2020) Appraising the genetic architecture of Kernel traits in hexaploid wheat using GWAS. Int J Mol Sci 21:1–21
Narum SR (2006) Beyond Bonferroni: less conservative analyses for conservation genetics. Conserv Genet 7:783–787
Nehe A, Akin B, Sanal T et al (2019) Genotype x environment interaction and genetic gain for grain yield and grain quality traits in Turkish spring wheat released between 1964 and 2010. PLoS ONE 14:1–18
Niu X, Chen S, Li J et al (2019) Genome-wide identification of GRAS genes in Brachypodium distachyon and functional characterization of BdSLR1 and BdSLRL1. BMC Genomics 20:1–18
Qaseem MF, Qureshi R, Muqaddasi QH et al (2018) Genome-wide association mapping in bread wheat subjected to independent and combined high temperature and drought stress. PLoS ONE 13:1–22
Rahimi Y, Bihamta MR, Taleei A et al (2019) Genome-wide association study of agronomic traits in bread wheat reveals novel putative alleles for future breeding programs. BMC Plant Biol 19:1–19
Ramya P, Chaubal A, Kulkarni K et al (2010) QTL mapping of 1000-kernel weight, kernel length, and kernel width in bread wheat (Triticum aestivum L.). J Appl Genet 51:421–429
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:1–8
Reif JC, Maurer HP, Korzun V et al (2011) Mapping QTLs with main and epistatic effects underlying grain yield and heading time in soft winter wheat. Theor Appl Genet 123:283–292
Richards RA (2000) Selectable traits to increase crop photosynthesis and yield of grain crops. J Exp Bot 51:447–458
Schilling S, Pan S, Kennedy A, Melzer R (2018) MADS-box genes and crop domestication: the jack of all traits. J Exp Bot 69:1447–1469
Sehgal D, Autrique E, Singh R et al (2017) Identification of genomic regions for grain yield and yield stability and their epistatic interactions. Sci Rep 7:1–12
Singh K, Batra R, Sharma S et al (2021) Wheat QTL db: A QTL database for wheat. Mol Genet Genomics (in press)
SPSS Inc (2008) Released, SPSS Statistics for Windows, Version 17.0. Chicago: SPSS Inc
Sun C, Dong Z, Zhao L et al (2020) The Wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol J 18:1354–1360
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Verslues PE, Zhu JK (2005) Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem Soc Trans 33:375–379
Walter S, Kahla A, Arunachalam C et al (2015) A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J Exp Bot 66:2583–2593
Wang RX, Hai L, Zhang XY et al (2009) QTL mapping for grain filling rate and yield-related traits in RILs of the Chinese winter wheat population Heshangmai x Yu8679. Theor Appl Genet 118:313–325
Wang S, Wong D, Forrest K et al (2014) Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wang SX, Zhu YL, Zhang DX et al (2017) Genome-wide association study for grain yield and related traits in elite wheat varieties and advanced lines using SNP markers. PLoS ONE 12:1–14
Ward BP, Brown-Guedira G, Kolb FL et al (2019) Genome-wide association studies for yield-related traits in soft red winter wheat grown in Virginia. PLoS ONE 14:1–28
White T, van der Ende J, Nichols TE (2019) Beyond Bonferroni revisited: concerns over inflated false positive research findings in the fields of conservation genetics, biology, and medicine. Conserv Genet 20:927–937
Wilson DJ (2019) The harmonic mean p-value for combining dependent tests. Proc Natl Acad Sci 116:1195–1200
Yadav AK, Carroll AJ, Estavillo GM et al (2019) Wheat drought tolerance in the field is predicted by amino acid responses to glasshouse-imposed drought. J Exp Bot 70:4931–4947
Yu LX, Lorenz A, Rutkoski J et al (2011) Association mapping and gene-gene interaction for stem rust resistance in CIMMYT spring wheat germplasm. Theor Appl Genet 123:1257–1268
Zhang Z, Ersoz E, Lai CQ et al (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355–360
Zhang YM, Jia Z, Dunwell JM (2019) Editorial: The applications of new multi-locus GWAS methodologies in the genetic dissection of complex traits. Front Plant Sci 10:1–6
Zhang L, Liu P, Wu J et al (2020) Identification of a novel ERF gene, TaERF8, associated with plant height and yield in wheat. BMC Plant Biol 20:1–12
Zhu T, Wang L, Rimbert H et al (2021) Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J 1–12
Zuka O, Schaffner SF, Samocha K et al (2014) Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci 111:E455–E464
Acknowledgements
The material for association panel and the genotypic data for this work was received from CIMMYT, Mexico. Thanks are due to the Department of Biotechnology (DBT), Govt of India for providing funds in the form of research projects awarded to Shailendra Sharma (SS). The authors are thankful to Ch. Charan Singh University, Meerut for providing laboratory and field facilities. HSB was awarded positions of INSA Senior Scientist/Honorary Scientist during the course of the study by INSA, New Delhi.
Author information
Authors and Affiliations
Contributions
SS (Shailendra Sharma) conceived and conducted or directed the experiments. PM, JK, SS1(SahadevSingh), SS2 (Shiveta Sharma) performed the experiments. JK, PKM, MS analysed the data. JKR, PKS, HSB, PKG and SS participated in manuscript writing. SS finalized the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consented to publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Malik, P., Kumar, J., Singh, S. et al. Single-trait, multi-locus and multi-trait GWAS using four different models for yield traits in bread wheat. Mol Breeding 41, 46 (2021). https://doi.org/10.1007/s11032-021-01240-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-021-01240-1