Abstract
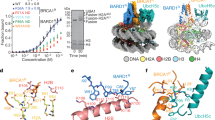
Protein ubiquitination at sites of DNA double-strand breaks (DSBs) by RNF168 recruits BRCA1 and 53BP11,2, which are mediators of the homologous recombination and non-homologous end joining DSB repair pathways, respectively3. Non-homologous end joining relies on 53BP1 binding directly to ubiquitinated lysine 15 on H2A-type histones (H2AK15ub)4,5 (which is an RNF168-dependent modification6), but how RNF168 promotes BRCA1 recruitment and function remains unclear. Here we identify a tandem BRCT-domain-associated ubiquitin-dependent recruitment motif (BUDR) in BRCA1-associated RING domain protein 1 (BARD1) (the obligate partner protein of BRCA1) that, by engaging H2AK15ub, recruits BRCA1 to DSBs. Disruption of the BUDR of BARD1 compromises homologous recombination and renders cells hypersensitive to PARP inhibition and cisplatin. We further show that BARD1 binds nucleosomes through multivalent interactions: coordinated binding of H2AK15ub and unmethylated H4 lysine 20 by its adjacent BUDR and ankyrin repeat domains, respectively, provides high-affinity recognition of DNA lesions in replicated chromatin and promotes the homologous recombination activities of the BRCA1–BARD1 complex. Finally, our genetic epistasis experiments confirm that the need for BARD1 chromatin-binding activities can be entirely relieved upon deletion of RNF168 or 53BP1. Thus, our results demonstrate that by sensing DNA-damage-dependent and post-replication histone post-translation modification states, BRCA1–BARD1 complexes coordinate the antagonization of the 53BP1 pathway with promotion of homologous recombination, establishing a simple paradigm for the governance of the choice of DSB repair pathway.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data are available in the Article and its Supplementary Information. Source data are provided with this paper.
References
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009).
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009).
Hustedt, N. & Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9 (2017).
Fradet-Turcotte, A. et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 (2013).
Wilson, M. D. et al. The structural basis of modified nucleosome recognition by 53BP1. Nature 536, 100–103 (2016).
Mattiroli, F. et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012).
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006).
Bothmer, A. et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell 42, 319–329 (2011).
Dimitrova, N., Chen, Y.-C. M., Spector, D. L. & de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 (2008).
Nakamura, K. et al. H4K20me0 recognition by BRCA1–BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 21, 311–318 (2019).
Manke, I. A., Lowery, D. M., Nguyen, A. & Yaffe, M. B. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302, 636–639 (2003).
Yu, X., Chini, C. C. S., He, M., Mer, G. & Chen, J. The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 (2003).
Glover, J. N. M., Williams, R. S. & Lee, M. S. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 29, 579–585 (2004).
Wu, Q., Jubb, H. & Blundell, T. L. Phosphopeptide interactions with BRCA1 BRCT domains: more than just a motif. Prog. Biophys. Mol. Biol. 117, 143–148 (2015).
Li, M. & Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 23, 693–704 (2013).
Billing, D. et al. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol. Cell 72, 127–139 (2018).
Natsume, T., Kiyomitsu, T., Saga, Y. & Kanemaki, M. T. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218 (2016).
Laufer, M. et al. Structural requirements for the BARD1 tumor suppressor in chromosomal stability and homology-directed DNA repair. J. Biol. Chem. 282, 34325–34333 (2007).
Birrane, G., Varma, A. K., Soni, A. & Ladias, J. A. A. Crystal structure of the BARD1 BRCT domains. Biochemistry 46, 7706–7712 (2007).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007).
Wang, B. et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 (2007).
Kim, H., Chen, J. & Yu, X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316, 1202–1205 (2007).
Sims, J. J. & Cohen, R. E. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell 33, 775–783 (2009).
Sato, Y. et al. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 (2009).
Hu, Y. et al. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 25, 685–700 (2011).
Shao, G. et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 23, 740–754 (2009).
Zgheib, O., Pataky, K., Brugger, J. & Halazonetis, T. D. An oligomerized 53BP1 Tudor domain suffices for recognition of DNA double-strand breaks. Mol. Cell. Biol. 29, 1050–1058 (2009).
Hu, Q., Botuyan, M. V., Cui, G., Zhao, D. & Mer, G. Mechanisms of ubiquitin-nucleosome recognition and regulation of 53BP1 chromatin recruitment by RNF168/169 and RAD18. Mol. Cell 66, 473–487 (2017).
Cao, J. & Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2, 26 (2012).
Densham, R. M. et al. Human BRCA1–BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 23, 647–655 (2016).
Kalb, R., Mallery, D. L., Larkin, C., Huang, J. T. & Hiom, K. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 8, 999–1005 (2014).
Witus, S. R. et al. BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1. Nat. Struct. Mol. Biol. 28, 268–277 (2021).
Chapman, J. R., Sossick, A. J., Boulton, S. J. & Jackson, S. P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 125, 3529–3534 (2012).
Pellegrino, S., Michelena, J., Teloni, F., Imhof, R. & Altmeyer, M. Replication-coupled dilution of H4K20me2 guides 53BP1 to pre-replicative chromatin. Cell Rep. 19, 1819–1831 (2017).
Nacson, J. et al. BRCA1 mutation-specific responses to 53BP1 loss-induced homologous recombination and PARP inhibitor resistance. Cell Rep. 24, 3513–3527 (2018).
Chen, J. et al. 53BP1 loss rescues embryonic lethality but not genomic instability of BRCA1 total knockout mice. Cell Death Differ. 27, 2552–2567 (2020).
Dyer, P. N. et al. in Methods in Enzymology Vol. 375 (eds Allis, C. D. & Wu, C.) 23–44 (Academic, 2003).
Simon, M. D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007).
Long, L., Furgason, M. & Yao, T. Generation of nonhydrolyzable ubiquitin-histone mimics. Methods 70, 134–138 (2014).
Wilson, M. D. et al. Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer. Nat. Commun. 10, 4189 (2019).
Acknowledgements
We thank all members of the laboratory of J.R.C. for discussions; D. Durocher for plasmid reagents; N. Johnson for discussions regarding unpublished data; and T. Humphrey for comments on the manuscript. This work was funded by a Cancer Research UK (CRUK) Career Development Fellowship (C52690/A19270 to J.R.C.), which previously provided salary support to J.R.B., and CRUK Oxford Centre funding (C5255/A18085 to J.R.B. and J.R.C.). J.R.B.’s salary is provided by a Ruth L. Kirschstein NRSA Individual Postdoctoral Fellowship (F32) (NIH/NCI - F32CA239339). The laboratory of J.R.C. is also supported by the Medical Research Council (MRC - MR/R017549/1), the MRC Molecular Haematology Unit (MRC MHU, UK) and the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC). J.R.C. holds a Lister Institute Research Prize Fellowship. The Wellcome Centre for Human Genetics (WHG) is supported by Wellcome core award 090532/Z/09/Z. C.B. was sponsored by an ERASMUS+ internship. M.D.W.’s work is supported by the Wellcome Trust (210493), the MRC (T029471/1) and the University of Edinburgh. The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust (203149). A.G.’s research is supported by the Lundbeck Foundation (R198-2015-269 and R313-2019-448), the European Research Council (ERC CoG no. 724436) and Independent Research Fund Denmark (7016-00042B and 4092-00404B). Research at CPR is supported by the Novo Nordisk Foundation (NNF14CC0001). We also acknowledge the BHF Centre of Research Excellence, Oxford (RE/13/1/30181), and the WHG Cellular Imaging core for equipment access and support.
Author information
Authors and Affiliations
Contributions
J.R.B. designed and analysed the majority of experiments, and supervised experiments performed by C.B. M.D.W. and G.C. undertook and analysed all in vitro nucleosome binding experiments. The project was initiated in collaboration with A.G. J.R.C. conceived and supervised the project, and designed and analysed experiments. J.R.B. and J.R.C. co-wrote the manuscript, with author input.
Corresponding author
Ethics declarations
Competing interests
A.G. is co-founder and CSO in Ankrin Therapeutics. No other authors have competing interests.
Additional information
Peer review information Nature thanks Daniel Durocher, Joanna Morris and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 BARD1 BRCT-mutated transgenes are stably expressed in BARD1AID/AID HCT-116 cells.
Related to Fig. 1. a, Immunoblots of whole-cell lysates collected at the indicated time points after IAA addition. Expression of the auxin-degron-targeting SCF-complex E3 ligase Oryza sativa TIR1 was induced using doxycycline (2 μg ml−1), 24 h before the depletion of endogenous BARD1–AID protein with IAA (1 mM). Representative of two biological repeats. b, Immunoblot from whole-cell lystes of BARD1 BRCT mutants screened for olaparib sensitivity. c, Immunoblot of whole cell lysates from BARD1AID/AID cells expressing the indicated transgenes. Cells were seeded in the presence of doxycycline (2 μg ml−1) and IAA (1 mM) was added after 24 h. Lysates were collected at the indicated time points after IAA addition. Representative of two biological repeats.
Extended Data Fig. 2 ARD and BRCTs in BARD1 cooperate in recruiting BRCA1 to post-replicative chromatin during homologous recombination.
Related to Fig. 2. a–c, Survival of the indicated BARD1AID/AID cell lines grown for 7 days without IAA in the presence of the indicated doses of olaparib or cisplatin. Cell lines were seeded in doxycycline (2 μg ml−1) for 24 h before olaparib or cisplatin addition. Resazurin cell viability assay, n = 3 biological experiments, mean ± s.d. d, High-content immunofluorescent microscopy of BRCA1 IRIF in H4K20me0+ BARD1AID/AID cells expressing the indicated transgenes. Cultures were grown in the presence of doxycycline (2 μg ml−1) for 24 h before IAA (1 mM) addition, irradiated 2 h later, and fixed following irradiation. Scale bar, 5 μm. Representative of n = 2 biological experiments. e, Top, quantification of BRCA1 IRIF from d. Boxes indicate the 25th–75th percentiles with the median denoted, and whiskers indicate the 10th–90th percentiles. BRCA1 foci measurements are made for nuclei in the bottom quartile of H4K20me0 integrated staining intensity (≥172 nuclei per condition). Foci quantification and H4K20me0 integrated intensity measurements were performed with CellProfiler. Bottom, mean number of BRCA1 foci per cell from two independent experiments ± s.d. f, g, Same as in d, e, respectively, in RAP80−/− cells. ≥ 179 nuclei per condition. h, i, Survival of the indicated BARD1AID/AID cell lines grown for 7 days without IAA in the presence of olaparib. Cell lines were seeded in doxycycline (2 μg ml−1) for 24 h before olaparib addition. Resazurin cell viability assay, n = 3 biological experiments, mean ± s.d
Extended Data Fig. 3 Purification of BARD1 and 53BP1 fragments and assembly of modified nucleosomes.
Related to Fig. 3. a, Western blot of HA-tagged 53BP1–BARD1 fusion proteins used in b stably expressed in BARD1AID/AID 53BP1−/− cells. b, Top, model depicting the 53BP1–BARD1 fusion protein. The fusion is a chimera composed of the 53BP1 minimal focus-forming region (amino acids 1220–1711) and BARD1 BRCTs (amino acids 555–777). Expressed form includes an N-terminal 2×HA–Flag epitope tag. Bottom, confocal immunofluorescent microscopy of 53BP1–BARD1 chimeric fusion proteins in irradiated BARD1AID/AID53BP1−/− cells. Cultures were grown in the presence of doxycycline (2 μg ml−1) for 24 h before IAA (1 mM) addition and irradiated (5 Gy) 2 h later. Cells were fixed 2 h following irradiation. Scale bar, 10 μm. Representative of n = 2 biological experiments. c, SDS–PAGE gel, stained with InstantBlue protein stain of proteins used in Fig. 3a. d, SDS–PAGE gel, stained with InstantBlue protein stain of nucleosomes used in this study. e, Native gel electrophoresis of Widom 601 DNA in isolation and wrapped with nucleosomes used in this study. f, Representative gel images from EMSA experiments quantified in Fig. 3b. H2AKc15ub-modified, or H2AKc15ub- and H4Kc20me2-modified, nucleosomes (or control DNA) were incubated with increasing concentrations of 6×His–MBP–BARD1(ARD–BRCT) or GST–53BP1(TTD–UDR). Complexes were resolved by native PAGE and visualized using Diamond DNA stain. g, SDS–PAGE gel, stained with InstantBlue protein stain of BARD1 variants used in Fig. 3c. Neighbouring lanes were loaded with two different concentrations.
Extended Data Fig. 4 BRCA1 recruitment to IRIF is RNF168-dependent, but independent of PRC1 and BRCA1–BARD1 ubiquitin ligase activity.
Related to Fig. 4. a, Model indicating the three major known sites of ubiquitin attachment on histone H2A and the genetic manipulations used to block each individually in our experiments. PRC1−/− indicates RING1A−/−RING1B−/− double knockout. b, Immunoblot of whole-cell lysates from BARD1AID/AID parental cells and RNF168−/− derivatives. Cells were seeded in the presence of doxycycline (2 μg ml−1) and IAA (1 mM) was added after 24 h. Lysates were collected 8 h after IAA addition. Representative of two biological repeats. c, Immunofluorescent microscopy of BARD1AID/AID parental cells and RNF168−/− derivatives. Cultures were seeded 24 h before irradiation (5 Gy), and fixed 2 h later. Representative of n = 3 biological experiments. Scale bar, 10 μm. d, Quantification of BRCA1 and 53BP1 IRIF from c. Boxes indicate the 25th–75th percentiles with the median denoted, and whiskers indicate the 10th–90th percentiles. BRCA1 foci measurements are made for EdU-positive nuclei. Foci quantification was performed using using CellProfiler. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. ****P ≤ 0.0001. Representative of n = 3 biological replicates. e, Immunoblot of whole-cell lysates from BARD1AID/AID parental cells and RING1A−/−, RING1B−/− and RING1A−/−RING1B−/− (denoted as PRC1−/−) derivatives. Cultures were seeded 24 h before irradiation (10 Gy), and collected 2 h later. f, g, The indicated cell lines were treated as in d. Representative of n = 3 biological replicates
Extended Data Fig. 5 The β2′–β3′ loop and ARD counteract toxic 53BP1-dependent non-homologous end joining.
Related to Fig. 4. a, Immunofluorescent microscopy of RAD51 IRIF in BARD1AID/AID cells expressing the indicated transgenes. Cultures were grown in the presence of doxycycline (2 μg ml−1) for 24 h before IAA (1 mM) addition, irradiated 2 h later (5 Gy), and fixed with PFA 2 h following irradiation. Data were collected from the same experiment as Fig. 2i, j. Scale bar, 5 μm. Representative of n = 3 biological experiments. b, Top, quantification of RAD51 foci per cell from a. Per condition, ≥255 nuclei. Boxes indicate the 25th–75th percentiles with the median denoted, and whiskers indicate the 10th–90th percentiles. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. ****P ≤ 0.0001, ***P = 0.0003. Data are from same experiment presented in Fig. 2i, j. BARD1AID/AID cells expressing GST and BARD1 are displayed in both for comparison. Representative of n = 3 biological experiments. Bottom, mean number of RAD51 IRIF from three independent biological experiments ± s.d. c, d, Immunoblot of whole-cell lysates from BARD1AID/AID (top) or BARD1AID/AID53BP1−/− (bottom) cells expressing the indicated transgenes. Cells were seeded in the presence of doxycycline (2 μg ml−1) and IAA (1 mM) was added after 24 h. Lysates were collected at the indicated time points after IAA addition. Representative of two biological repeats. e–h, Survival of the indicated BARD1AID/AID cell lines grown without IAA for 7 days in the presence of the indicated doses of olaparib or cisplatin. Cultures were seeded in doxycycline (2 μg ml−1) and olaparib or cisplatin was added 24 h later. Survival was measured after 7 days by resazurin cell viability assay (n = 3 biological experiments) mean ± s.d
Supplementary information
Supplementary Figure
This file contains the uncropped blots shown in Fig. 3 and Extended Data Figs 1, 3, 4 and 5.
Rights and permissions
About this article
Cite this article
Becker, J.R., Clifford, G., Bonnet, C. et al. BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination. Nature 596, 433–437 (2021). https://doi.org/10.1038/s41586-021-03776-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03776-w
This article is cited by
-
DNMT3B PWWP mutations cause hypermethylation of heterochromatin
EMBO Reports (2024)
-
Recent advances in chemical protein synthesis: method developments and biological applications
Science China Chemistry (2024)
-
Genetic separation of Brca1 functions reveal mutation-dependent Polθ vulnerabilities
Nature Communications (2023)
-
Mechanisms of synthetic lethality between BRCA1/2 and 53BP1 deficiencies and DNA polymerase theta targeting
Nature Communications (2023)
-
An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.