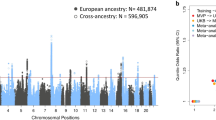
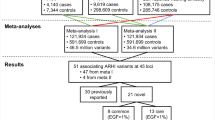
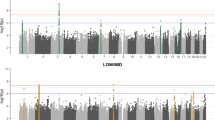
Abstract
Genome-wide association studies (GWAS) provide an unbiased first look at genetic loci involved in aging and noise-induced sensorineural hearing loss and tinnitus. The hearing phenotype, whether audiogram-based or self-report, is regressed against genotyped information at representative single nucleotide polymorphisms (SNPs) across the genome. Findings include the fact that both hearing loss and tinnitus are polygenic disorders, with up to thousands of genes, each of effect size of < 0.02. Smaller human GWAS’ were able to use objective measures and identified a few loci; however, hundreds of thousands of participants have been required for the statistical power to identify significant variants, and GWAS is unable to assess rare variants with mean allele frequency < 1%. Animal studies are required as well because of inability to access the human cochlea. Mouse GWAS builds on linkage techniques and the known phenotypic differences in auditory function between inbred strains. With the advantage that the laboratory environment can be controlled for noise and aging, the Hybrid Mouse Diversity Panel (HDMP) combines 100 strains sequenced at high resolution. Lift-over regions between mice and humans have identified over 17,000 homologous genes. Since most significant SNPs are either intergenic or in introns, and binding sites between species are poorly preserved between species, expression quantitative trait locus information is required to bring humans and mice into agreement. Transcriptome-wide analysis studies (TWAS) can prioritize putative causal genes and tissues. Diverse species, each making a distinct contribution, carry a synergistic advantage in the quest for treatment and ultimate cure of sensorineural hearing difficulties.



Similar content being viewed by others
Material availability
Not applicable.
References
Ahmadmehrabi S, Li B, Park J et al (2021) Genome-first approach to rare EYA4 variants and cardio-auditory phenotypes in adults. Hum Genet 140:957–967. https://doi.org/10.1007/s00439-021-02263-6
Amanat S, Requena T, Lopez-Escamez JA (2020) A Systematic review of extreme phenotype strategies to search for rare variants in genetic studies of complex disorders. Genes. https://doi.org/10.3390/genes11090987
Amanat S, Gallego-Martinez A, Sollini J et al (2021) Burden of rare variants in synaptic genes in patients with severe tinnitus: An exome based extreme phenotype study. EBioMedicine. https://doi.org/10.1016/j.ebiom.2021.103309
Anwar MN, Oakes MP (2012) Data mining of audiology patient records: Factors influencing the choice of hearing aid type. BMC Med Inform Decis Mak 12:1–8. https://doi.org/10.1186/1472-6947-12-S1-S6
Bennett BJ, Farber CR, Orozco L et al (2010) A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. https://doi.org/10.1101/gr.099234.109
Bogo R, Farah A, Karlsson KK et al (2017) Prevalence, incidence proportion, and heritability for tinnitus. Ear Hear 38:292–300. https://doi.org/10.1097/AUD.0000000000000397
Bolajoko O, Davis AC, Hoffman HJ (2019) Hearing loss: Rising prevalence and impact. Bull World Health Organ 97:646-646A. https://doi.org/10.2471/BLT.19.224683
Boussaty EC, Gillard D, Lavinsky J et al (2020) The Genetics of variation of the wave 1 amplitude of the mouse auditory brainstem response. J Assoc Res Otolaryngol. https://doi.org/10.1007/s10162-020-00762-3
Breschi A, Gingeras TR, Guigó R (2017) Comparative transcriptomics in human and mouse. Nat Rev Genet 18:425–440. https://doi.org/10.1038/nrg.2017.19
Brubaker DK, Proctor EA, Haigis KM, Lauffenburger DA (2019) Computational translation of genomic responses from experimental model systems to humans. PLoS Comput Biol 15:e1006286. https://doi.org/10.1371/journal.pcbi.1006286
Bycroft C, Freeman C, Petkova D et al (2018) Genome-wide genetic data on 500,000 UK Biobank participants. Nature 562:203–209. https://doi.org/10.1038/s41586-018-0579-z
Cederroth CR, Pirouzifard M, Trpchevska N et al (2019) Association of genetic vs environmental factors in swedish adoptees with clinically significant tinnitus. JAMA Otolaryngol Head Neck Surg 145:222–229. https://doi.org/10.1001/jamaoto.2018.3852
Chatzinakos C, Georgiadis F, Lee D et al (2020) TWAS pathway method greatly enhances the number of leads for uncovering the molecular underpinnings of psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. https://doi.org/10.1002/ajmg.b.32823
Cherny SS, Livshits G, Wells HRR et al (2020) Self-reported hearing loss questions provide a good measure for genetic studies: a polygenic risk score analysis from UK Biobank. Eur J Hum Genet 28:1056–1065. https://doi.org/10.1038/s41431-020-0603-2
Chia R, Achilli F, Festing MFW, Fisher EMC (2005) The origins and uses of mouse outbred stocks. Nat Genet. https://doi.org/10.1038/ng1665
Clifford RE, Maihofer AX, Stein MB et al (2020) Novel risk loci in tinnitus and causal inference with neuropsychiatric disorders among adults of european ancestry. JAMA Otolaryngol Head Neck Surg 146:1015–1025. https://doi.org/10.1001/jamaoto.2020.2920
Crow AL, Ohmen J, Wang J et al (2015) The genetic architecture of hearing impairment in mice: evidence for frequency-specific genetic determinants. G3 (Bethesda). https://doi.org/10.1534/g3.115.021592
Demeester K, van Wieringen A, Hendrickx J et al (2010) Heritability of audiometric shape parameters and familial aggregation of presbycusis in an elderly Flemish population. Hear Res 265:1–10. https://doi.org/10.1016/j.heares.2010.03.007
Demopoulos C, Duong X, Hinkley LB et al (2020) Global resting-state functional connectivity of neural oscillations in tinnitus with and without hearing loss. Hum Brain Mapp 41:2846–2861. https://doi.org/10.1002/hbm.24981
DeStefano AL, Gates GA, Heard-Costa N et al (2003) Genome-wide linkage analysis to presbycusis in the Framingham Heart Study. Arch Otolaryngol Head Neck Surg 129:285–289. https://doi.org/10.1001/archotol.129.3.285
Drayton M, Noben-Trauth K (2006) Mapping quantitative trait loci for hearing loss in Black Swiss mice. Hear Res. https://doi.org/10.1016/j.heares.2005.11.006
Elgoyhen AB, Langguth B, De Ridder D, Vanneste S (2015) Tinnitus: perspectives from human neuroimaging. Nat Rev Neurosci 16:632–642. https://doi.org/10.1038/nrn4003
Flint J, Eskin E (2012) Genome-wide association studies in mice. Nat Rev Genet 13:807–817. https://doi.org/10.1038/nrg3335
Fransen E, Lemkens N, Van Laer L, Van Camp G (2003) Age-related hearing impairment (ARHI): environmental risk factors and genetic prospects. Exp Gerontol 38:353–359. https://doi.org/10.1016/S0531-5565(03)0032-9
Friedman RA, van Laer L, Huentelman MJ et al (2009) GRM7 variants confer susceptibility to age-related hearing impairment. Hum Mol Genet 18:785–796. https://doi.org/10.1093/hmg/ddn402
Gabriela S, Sanches G, Samelli AG, Nishiyama AK (2010) GIN Test (Gaps-in-Noise) in normal listeners with and without tinnitus. SciElo Brasil 22:257–262. https://doi.org/10.1590/S0104-56872010000300017
Garringer HJ, Pankratz ND, Nichols WC, Reed T (2006) Hearing impairment susceptibility in elderly men and the DFNA18 locus. Arch Otolaryngol Head Neck Surg 132:506–510. https://doi.org/10.1001/archotol.132.5.506
Gaziano JM, Concato J, Brophy M et al (2016) Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70:214–223. https://doi.org/10.1016/j.jclinepi.2015.09.016
Ghazalpour A, Rau CD, Farber CR et al (2012) Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm Genome. https://doi.org/10.1007/s00335-012-9411-5
Gilles A, Schlee W, Rabau S et al (2016) Decreased speech-in-noise understanding in young adults with tinnitus. Front Neurosci 10:1–14. https://doi.org/10.3389/fnins.2016.00288
Gilles A, van Camp G, van De Heyning P, Fransen E (2017) A pilot genome-wide association study identifies potential metabolic pathways involved in tinnitus. Front Neurosci 11:1–10. https://doi.org/10.3389/fnins.2017.00071
Girotto G, Pirastu N, Sorice R et al (2011) Hearing function and thresholds: A genome-wide association study in European isolated populations identifies new loci and pathways. J Med Genet 48:369–374. https://doi.org/10.1136/jmg.2010.088310
Heffner HE, Heffner RS (2012) Behavioral tests for tinnitus in animals, 1st edn. Springer Science and Business media, New York, p 21–58
Henry KR (1982) Influence of genotype and age on noise-induced auditory losses. Behav Genet. https://doi.org/10.1007/BF01070410
Herzig AF, Nutile T, Babron MC et al (2018) Strategies for phasing and imputation in a population isolate. Genet Epidemiol. https://doi.org/10.1002/gepi.22109
Hoffmann TJ, Keats BJ, Yoshikawa N et al (2016) A Large genome-wide association study of age-related hearing impairment using electronic health records. PLoS Genet 12:1–20. https://doi.org/10.1371/journal.pgen.1006371
Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. https://doi.org/10.1371/journal.pgen.1000529
Huyghe JR, van Laer L, Hendrickx JJ et al (2008) Genome-wide SNP-based linkage scan identifies a locus on 8q24 for an age-related hearing impairment trait. Am J Hum Genet 83:401–407. https://doi.org/10.1016/j.ajhg.2008.08.002
Johnson K, Zheng Q (2002) Ahl2, a second locus affecting age-related hearing loss in mice. Genomics 80:461–464
Johnson KR, Erway LC, Cook SA et al (1997) A major gene affecting age-related hearing loss in C57BL/6J mice. Hear Res. https://doi.org/10.1016/S0378-5955(97)00155-X
Johnson KR, Zheng QY, Noben-Trauth K (2006) Strain background effects and genetic modifiers of hearing in mice. Brain Res. https://doi.org/10.1016/j.brainres.2006.02.021
Johnson KR, Longo-Guess C, Gagnon LH et al (2008) A locus on distal chromosome 11 (ahl8) and its interaction with Cdh23ahl underlie the early onset, age-related hearing loss of DBA/2J mice. Genomics. https://doi.org/10.1016/j.ygeno.2008.06.007
Kalra G, Milon B, Casella AM et al (2020) Biological insights from multi-omic analysis of 31 genomic risk loci for adult hearing difficulty. PLoS Genet 16:1–32. https://doi.org/10.1371/journal.pgen.1009025
Kang HM, Zaitlen NA, Wade CM et al (2008) Efficient control of population structure in model organism association mapping. Genetics. https://doi.org/10.1534/genetics.107.080101
Karlsson KK, Harris JR, Svartengren M (1997) Description and primary results from an audiometric study of male twins. Ear Hear 18(2). https://journals.lww.com/ear-hearing/Fulltext/1997/04000/Description_and_Primary_Results_from_an.3.aspx
Kvestad E, Czajkowski N, Krog NH et al (2012) Heritability of hearing loss. Epidemiology 23:328–331. https://doi.org/10.1097/EDE.0b013e318245996e
Latoche JR, Neely HR, Noben-Trauth K (2011) Polygenic inheritance of sensorineural hearing loss (Snhl2, -3, and -4) and organ of Corti patterning defect in the ALR/LtJ mouse strain. Hear Res. https://doi.org/10.1016/j.heares.2010.12.017
Lavinsky J, Crow AL, Pan C et al (2015) Genome-wide association study identifies nox3 as a critical gene for susceptibility to noise-induced hearing loss. PLoS Genet 11:e1005094. https://doi.org/10.1371/journal.pgen.1005094
Lavinsky J, Ge M, Crow AL et al (2016) The genetic architecture of noise-induced hearing loss: evidence for a gene-by-environment interaction. G3 (Bethesda). https://doi.org/10.1534/g3.116.032516
Leaver AM, Seydell-Greenwald A, Turesky TK et al (2012) Cortico-limbic morphology separates tinnitus from tinnitus distress. Front Syst Neurosci 6:21. https://doi.org/10.3389/fnsys.2012.00021
Lewis MA, Nolan LS, Cadge BA et al (2018) Whole exome sequencing in adult-onset hearing loss reveals a high load of predicted pathogenic variants in known deafness-associated genes and identifies new candidate genes. BMC Med Genom 11:1–12. https://doi.org/10.1186/s12920-018-0395-1
Liberman MC, Kujawa SG (2017) Cochlear synaptopathy in acquired sensorineural hearing loss: Manifestations and mechanisms. Hear Res 349:138–147. https://doi.org/10.1016/j.heares.2017.01.003
Liberman MC, Epstein MJ, Cleveland SS et al (2016) Toward a differential diagnosis of hidden hearing loss in humans. PLoS ONE 11:1–15. https://doi.org/10.1371/journal.pone.0162726
Lin X, Chen Y, Wang M et al (2020) Altered topological patterns of gray matter networks in tinnitus: a graph-theoretical-based study. Front Neurosci 14:1–12. https://doi.org/10.3389/fnins.2020.00541
Lindblad-Toh K, Winchester E, Daly MJ et al (2000) Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat Genet. https://doi.org/10.1038/74215
Lusis AJ, Seldin MM, Allayee H et al (2016) The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. J Lipid Res. https://doi.org/10.1194/jlr.R066944
Maas IL, Brüggemann P, Requena T et al (2017) Genetic susceptibility to bilateral tinnitus in a Swedish twin cohort. Genet Med. https://doi.org/10.1038/gim.2017.4
Mashimo T, Erven AE, Spiden SL et al (2006) Two quantitative trait loci affecting progressive hearing loss in 101/H mice. Mamm Genome. https://doi.org/10.1007/s00335-004-2438-5
Maynard RD, Ackert-Bicknell CL (2019) Mouse models and online resources for functional analysis of osteoporosis genome-wide association studies. Front Endocrinol. https://doi.org/10.3389/fendo.2019.00277
McCullagh M, Raymond D, Kerr M, Lusk S (2011) Prevalence of hearing loss and accuracy of self-report among factory workers. Noise Health 13:340–347
Momi SK, Wolber LE, Fabiane SM et al (2015) Genetic and environmental factors in age-related hearing impairment. Twin Res Hum Genet 18:383–392. https://doi.org/10.1017/thg.2015.35
Nagtegaal AP, Broer L, Zilhao NR et al (2019) Genome-wide association meta-analysis identifies five novel loci for age-related hearing impairment. Sci Rep 9:1–10. https://doi.org/10.1038/s41598-019-51630-x
Nemoto M, Morita Y, Mishima Y et al (2004) Ahl3, a third locus on mouse chromosome 17 affecting age-related hearing loss. Biochem Biophys Res Commun. https://doi.org/10.1016/j.bbrc.2004.09.186
Newman D, Fisher L, Ohmen J et al (2008) GRM7 variants associated with age-related hearing loss based on auditory perception. Hear Res 64:2391–2404. https://doi.org/10.1038/jid.2014.371
Niu Y, Xie C, Du Z et al (2021) Genome-wide association study identifies 7q11.22 and 7q36.3 associated with noise-induced hearing loss among Chinese population. J Cell Mol Med 25:411–420. https://doi.org/10.1111/jcmm.16094
Noben-Trauth K, Zheng QY, Johnson KR (2003) Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat Genet. https://doi.org/10.1038/ng1226
Noben-Trauth K, Latoche JR, Neely HR, Bennett B (2010) Phenotype and Genetics of Progressive Sensorineural Hearing Loss (Snhl1) in the LXS Set of Recombinant Inbred Strains of Mice. PLoS ONE. https://doi.org/10.1371/journal.pone.0011459
Normand R, Du W, Briller M, Gaujoux R, Starosvetsky E, Ziv-Kenet A, Shalev-Malul G, Tibshirani RJ, Shen-Orr SS (2018) Found in translation: a machine learning model for mouse-to-human inference. Nat Methods 18:1067–107.3 https://doi.org/10.1038/s41592-018-0214-9
Ohlemiller KK, Kiener AL, Gagnon PM (2016) QTL Mapping of endocochlear potential differences between C57BL/6J and BALB/cJ mice. J Assoc Res Otolaryngol. https://doi.org/10.1007/s10162-016-0558-8
Ohmen J, Kang EY, Li X et al (2014) Genome-wide association study for age-related hearing loss (AHL) in the mouse: a meta-analysis. J Assoc Res Otolaryngol 15:335–352. https://doi.org/10.1007/s10162-014-0443-2
Pastina MM, Pinto LR, Oliveira KM et al (2019) Opportunities and chanllenges of TWAS. Nat Genet 51:117–148. https://doi.org/10.1038/s41588-019-0385-z
Prewitt A, Harker G, Gilbert TA et al (2021) Mental health symptoms among Veteran va users by tinnitus severity: a population-based survey. Mil Med 186:167–175. https://doi.org/10.1093/milmed/usaa288
Rabinowitz PM, Galusha D, Slade MD et al (2006) Audiogram notches in noise-exposed workers. Ear Hear 27:742–750. https://doi.org/10.1097/01.aud.0000240544.79254.bc
Rentzsch P, Witten D, Cooper GM et al (2019) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. https://doi.org/10.1093/nar/gky1016
Salehi P, Ge MX, Gundimeda U et al (2017) Role of Neuropilin-1/Semaphorin-3A signaling in the functional and morphological integrity of the cochlea. PLoS Genet. https://doi.org/10.1371/journal.pgen.1007048
Shin J-B, Longo-Guess CM, Gagnon LH et al (2010) The R109H Variant of Fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of DBA/2J Mice. J Neurosci. https://doi.org/10.1523/JNEUROSCI.1541-10.2010
Smith LI (2002) A tutorial on principal components analysis introduction. http://www.cs.otago.ac.nz/cosc453/student_tutorials/principal_components.pdf
Tegg-Quinn S, Bennett RJ, Eikelboom RH, Baguley DM (2016) The impact of tinnitus upon cognition in adults: A systematic review. Int J Audiol 55:533–540. https://doi.org/10.1080/14992027.2016.1185168
van Laer L, Huyghe J, Hannula S et al (2010) A genome-wide association study for age-related hearing impairment in the Saami. Eur J Hum Genet 18:685–693
Visscher PM, McCarthy M, Yang J (2012) Five years of GWAS discovery. Am J Hum Genet 90:7–24. https://doi.org/10.1016/j.ajhg.2011.11.029
Visscher PM, Wray NR, Zhang Q et al (2017) 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet 101:5–22. https://doi.org/10.1016/j.ajhg.2017.06.005
Vuckovic D, Dawson S, Scheffer DI et al (2015) Genome-wide association analysis on normal hearing function identifies PCDH20 and SLC28A3 as candidates for hearing function and loss. Hum Mol Genet 24:5655–5664. https://doi.org/10.1093/hmg/ddv279
Vuckovic D, Mezzavilla M, Cocca M et al (2018) Whole-genome sequencing reveals new insights into age-related hearing loss: cumulative effects, pleiotropy and the role of selection. Eur J Hum Genet 26:1167–1179. https://doi.org/10.1038/s41431-018-0126-2
Wells HRR, Freidin MB, Abidin FNZ et al (2019) Genome-wide association study identifies 44 independent genomic loci for self-reported adult hearing difficulty in the UK Biobank cohort. Am J Hum Genet. https://doi.org/10.1101/549071
Wells H, Abidin F, Freidin M et al (2020) Genetic variation in RCOR1 is associated with tinnitus in UK Biobank. Bioreview Sept 24:1–20
White CH, Ohmen JD, Sheth S et al (2009) Genome-wide screening for genetic loci associated with noise- induced hearing loss. Mamm Genome 20:207–213. https://doi.org/10.1007/s00335-009-9178-5
Wiltshire T, Pletcher MT, Batalov S et al (2003) Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.0130101100
Wingfield A, Panizzon M, Grant MDM et al (2007) A twin-study of genetic contributions to hearing acuity in late middle age. J Gerontol A Biol Sci Med Sci 62:1294–1299. https://doi.org/10.1111/j.1365-2958.2011.07804.x
Yalcin B, Fullerton J, Miller S et al (2004) Unexpected complexity in the haplotypes of commonly used inbred strains of laboratory mice. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.0401189101
Zheng QY, Johnson KR, Erway LC (1999) Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res. https://doi.org/10.1016/S0378-5955(99)00003-9
Zheng QY, Ding D, Yu H et al (2009) A locus on distal chromosome 10 (ahl4) affecting age-related hearing loss in A/J mice. Neurobiol Aging. https://doi.org/10.1016/j.neurobiolaging.2007.12.011
Acknowledgements
This study was supported by NIDCD Grant 1R01DC018566-01A1. The authors are grateful to the Mills Auditory Foundation. We thank all the participants in the United Kingdom Biobank and the Veterans Administration Million Veteran Program.
Funding
This study was supported by Grant 1R01DC018566-01A1 of National Institutes on Deafness and Other Communications Disorders (NIDCD).
Author information
Authors and Affiliations
Consortia
Contributions
This was an invited article. Dr. Ely Boussaty performed literature search on mouse genomics and Dr. Clifford performed the literature search on human GWAS. Each drafted his/her work. All authors critically revised and approved of the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethics approval
Not applicable.
Consent
Not applicable.
Data
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Boussaty, E.C., Friedman, R.A., Million Veteran Program. et al. Hearing loss and tinnitus: association studies for complex-hearing disorders in mouse and man. Hum Genet 141, 981–990 (2022). https://doi.org/10.1007/s00439-021-02317-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02317-9