Abstract
Programmed cell death (PCD) in dinoflagellates has been introduced as a new concept that facilitates the demise of harmful algal blooms. Metacaspases (MCAs) play a role in PCD, but their function in dinoflagellates is unclear. Here, we cloned a novel MCA gene (PmMCA) from the harmful dinoflagellate Prorocentrum minimum and examined its molecular characteristics and gene expression during cell death. The gene was encoded in the nuclear genome with two introns. The putative protein contained 288 amino acids and three conserved MCA signature motifs. Phylogenetic analysis showed that PmMCA may have the same ancestor as other dinoflagellates. PmMCA expression and cell apoptosis were significantly induced under copper exposure, considerably affecting cell growth. These results suggest that PmMCA could be involved in PCD triggered by copper stress.




Similar content being viewed by others
Abbreviations
- aa:
-
Amino acids
- BLAST:
-
Basic local alignment search tool
- CTAB:
-
Cetyltrimethylammonium bromide
- EC50 :
-
Median effective concentration
- EST:
-
Expressed sequence tag
- HABs:
-
Harmful algal blooms
- MCA:
-
Metacaspase
- M13F:
-
M13 forward
- M13R:
-
M13 reverse
- NJ:
-
Neighbor-joining
- ORF:
-
Open reading frame
- PCD:
-
Programmed cell death
- PI:
-
Propidium iodide
- Pm:
-
Prorocentrum minimum
- RACE:
-
Rapid amplification of cDNA ends
- UTR:
-
Untranslated region
- DinoSL:
-
Dinoflagellate spliced leader
References
Taylor FJR, Hoppenrath M, Saldarriaga JF (2008) Dinoflagellate diversity and distribution. Biodivers Conserv 17:407–418. https://doi.org/10.1007/s10531-007-9258-3
Kudela RM, Gobler CJ (2012) Harmful dinoflagellate blooms caused by Cochlodinium sp.: global expansion and ecological strategies facilitating bloom formation. Harmful Algae 14:71–86. https://doi.org/10.1016/j.hal.2011.10.015
Huang K, Feng Q, Zhang Y, Ou L, Cen J, Lu S, Qi Y (2020) Comparative uptake and assimilation of nitrate, ammonium, and urea by dinoflagellate Karenia mikimotoi and diatom Skeletonema costatum s.l. in the coastal waters of the East China Sea. Mar Pollut Bull 155:111200. https://doi.org/10.1016/j.marpolbul.2020.111200
Spungin D, Bidle KD, Berman-Frank I (2019) Metacaspase involvement in programmed cell death of the marine cyanobacterium Trichodesmium. Environ Microbiol 21:667–681. https://doi.org/10.1111/1462-2920.14512
Franklin DJ, Brussaard CPD, Berges JA (2006) What is the role and nature of programmed cell death in phytoplankton ecology? Eur J Phycol 41:1–14. https://doi.org/10.1080/09670260500505433
Johnson JG, Janech MG, Van Dolah FM (2014) Caspase-like activity during aging and cell death in the toxic dinoflagellate Karenia brevis. Harmful Algae 31:41–53. https://doi.org/10.1016/j.hal.2013.08.005
Vavilala SL, Gawde KK, Sinha M, Souza JS, European JD (2015) Programmed cell death is induced by hydrogen peroxide by not by excessive ionic stress of sodium chloride in the unicellular green alga Chlamydomonas reinhardtii. Eur J Phycol 50:422–438. https://doi.org/10.1080/09670262.2015.1070437
Rogers HJ (2005) Cell death and organ development in plants. Curr Top Dev Biol 71:225–261. https://doi.org/10.1016/S0070-2153(05)71007-3
Pokrzywinski KL, Tilney CL, Warner ME, Coyne KJ (2017) Cell cycle arrest and biochemical changes accompanying cell death in harmful dinoflagellates following exposure to bacterial algicide IRI-160AA. Sci Rep 7:45102. https://doi.org/10.1038/srep45102
Locato V, De Gara L (2018) Programmed cell death in plants: an overview. Methods Mol Biol 1743:1–8. https://doi.org/10.1007/978-1-4939-7668-3_1
Choi CJ, Berges JA (2013) New types of metacaspases in phytoplankton reveal diverse origins of cell death proteases. Cell Death Dis 4:e490. https://doi.org/10.1038/cddis.2013.21
Asplund-Samuelsson J, Sundh J, Dupont CL, Allen AE, McCrow JP, Celepli NA, Bergman B, Ininbergs K, Ekman M (2016) Diversity and expression of bacterial metacaspases in an aquatic ecosystem. Front Microbiol 7:1043. https://doi.org/10.3389/fmicb.2016.01043
Bidle KD, Bender SJ (2008) Iron starvation and culture age activate metacaspases and programmed cell death in the marine diatom Thalassiosira pseudonana. Eukaryot Cell 7:223–236. https://doi.org/10.1128/EC.00296-07
Thamatrakoln K, Korenovska O, Niheu AK, Bidle KD (2012) Whole-genome expression analysis reveals a role for death-related genes in stress acclimation of the diatom Thalassiosira pseudonana. Environ Microbiol 14:67–81. https://doi.org/10.1111/j.1462-2920.2011.02468.x
Zhang X, Yang G, Liu Y, Yu W, Pan K, Li RX, Zhu M (2006) Induction of programed cell death in aging Prorocentrum donghaiense cells as was evidenced preliminarily by the identification of associated transcripts. Acta Biol Hung 57:473–483. https://doi.org/10.1556/ABiol.57.2006.4.9
Aranda M, Li Y, Liew YJ, Baumgarten S, Simakov O, Wilson MC, Piel J, Ashoor H, Bougouffa S, Bajic VB, Ryu T, Ravasi T, Bayer T, Micklem G, Kim H, Bhak J, LaJeunesse TC, Voolstra CR (2016) Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci Rep 6:39734. https://doi.org/10.1038/srep39734
Wang H, Park BS, Lim W-A, Ki J-S (2018) CpMCA, a novel metacaspase gene from the harmful dinoflagellate Cochlodinium polykrikoides and its expression during cell death. Gene 651:70–78. https://doi.org/10.1016/j.gene.2018.02.002
Okamoto OK, Hastings JW (2003) Genome-wide analysis of redox-regulated genes in a dinoflagellate. Gene 321:73–81. https://doi.org/10.1016/j.gene.2003.07.003
Guo R, Wang H, Suh YS, Ki J-S (2016) Transcriptomic profiles reveal the genome-wide responses of the harmful dinoflagellate Cochlodinium polykrikoides when exposed to the algicide copper sulfate. BMC Genomics 17:29. https://doi.org/10.1186/s12864-015-2341-3
Wang H, Abassi S, Ki J-S (2019) Origin and roles of a novel copper-zinc superoxide dismutase gene from the harmful dinoflagellate Prorocentrum minimum. Gene 683:113–122. https://doi.org/10.1016/j.gene.2018.10.013
Guo R, Ebenezer V, Ki J-S (2012) Transcriptional responses of heat shock protein 70 (HSP70) to thermal, bisphenol A, and copper stresses in the dinoflagellate Prorocentrum minimum. Chemosphere 89:512–520. https://doi.org/10.1016/j.chemosphere.2012.05.014
Wang H, Kim H, Lim W-A, Ki J-S (2019) Molecular cloning and oxidative-stress responses of a novel manganese superoxide dismutase (MnSOD) gene in the dinoflagellate Prorocentrum minimum. Mol Biol Rep 46:5955–5966. https://doi.org/10.1007/s11033-019-05029-6
Heil CA, Glibert PM, Fan CL (2005) Prorocentrum minimum (Pavillard) Schiller: a review of a harmful algal bloom species of growing worldwide importance. Harmful Algae 4:449–470. https://doi.org/10.1016/j.hal.2004.08.003
Stein JR (1973) Handbook of phycological methods: culture methods and growth measurements. Cambridge University Press, London, p 448
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326. https://doi.org/10.1093/nar/8.19.4321
Maruyama IN, Rakow TL, Maruyama HI (1995) cRACE: a simple method for identification of the 5′end of mRNAs. Nucleic Acids Res 23:3796–3797. https://doi.org/10.1093/nar/23.18.3796
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Wang H, Kim H, Ki J-S (2021) Transcriptomic identification and expression analysis of cold shock domain protein (CSP) genes in the marine dinoflagellate Prorocentrum minimum. J Appl Phycol 33:843–854. https://doi.org/10.1007/s10811-020-02332-9
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆Ct method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Zhang H, Hou Y, Miranda L, Campbell DA, Sturm NR, Gaasterland T, Lin S (2007) Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci USA 104:4618–4623. https://doi.org/10.1073/pnas.0700258104
Guo R, Ki J-S (2011) Spliced leader sequences detected in EST data of the dinoflagellates Cochlodinium polykrikoides and Prorocentrum minimum. Algae 26:229–235. https://doi.org/10.4490/algae.2011.26.3.229
Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inzé D, Van Breusegem F (2004) Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem 279:45329–45336. https://doi.org/10.1074/jbc.M406329200
Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam E, Bozhkov PV (2011) Metacaspases. Cell Death Differ 18:1279–1288. https://doi.org/10.1038/cdd.2011.66
Carvalho RN, Bopp SK, Lettieri T (2011) Transcriptomics responses in marine diatom Thalassiosira pseudonana exposed to the polycyclic aromatic hydrocarbon benzo[a]pyrene. PLoS ONE 6:e26985. https://doi.org/10.1371/journal.pone.0026985
Kebeish R, El-Ayouty Y, Husain A (2014) Effect of copper on growth, bioactive metabolites, antioxidant enzymes and photosynthesis-related gene transcription in Chlorella vulgaris. World J Biol Biol Sci 2:34–43
Abassi S, Wang H, Ponmani T, Ki J-S (2019) Small heat shock protein genes of the green algae Closterium ehrenbergii: Cloning and differential expression under heat and heavy metal stresses. Environ Toxicol 34:1013–1024. https://doi.org/10.1002/tox.22772
Ebenezer V, Lim W-A, Ki J-S (2014) Effects of the algicides CuSO4 and NaOCl on various physiological parameters in the harmful dinoflagellate Cochlodinium polykrikoides. J Appl Phycol 26:2357–2365. https://doi.org/10.1007/s10811-014-0267-9
Segovia M, Berges JA (2009) Inhibition of caspase-like activities prevents the appearance of reactive oxygen species and dark-induced apoptosis in the unicellular chlorophyte Dunaliella tertiolecta. J Phycol 45:1116–1126. https://doi.org/10.1111/j.1529-8817.2009.00733.x
Jauzein C, Erdner DL (2013) Stress-related responses in Alexandrium tamarense cells exposed to environmental changes. J Eukaryot Microbiol 60:526–538. https://doi.org/10.1111/jeu.12065
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIT) (No. 2020R1A2C2013373), and a part of the project titled ‘Development of hull adherent organism management technology (20210651),’ funded by the Ministry of Oceans and Fisheries, Korea.
Author information
Authors and Affiliations
Contributions
HW performed the study and drafted the manuscript; JSK revised and edited the manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2021_2617_MOESM1_ESM.tif
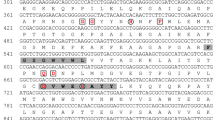
Supplementary file1 Multiple sequence alignments (A) and homoly matrix (B) of deduced amino acid sequences of PmMCA with those of other MCAs. Amino acid sequences from 10 MCAs share similar residues. A short linker was present between P20 and P10 subunits. The signature motifs were marked with black box. GenBank accession numbers of aligned proteins are as follows: Margalefidinium polykrikoides, AVD29967; Microcystis aeruginosa, WP_002768321; Gregarina niphandrodes, XP_011130889; Fistulifera solaris, GAX24014; Coccomyxa subellipsoidea, XP_005649943; Gonium pectorale, KXZ52917; Erythranthe guttata, XP_012858139; Zostera marina, KMZ57843; Prorocentrum micans, comp12808_c0 was obtained from P. micans transcriptome data. (TIF 2500 kb)
284_2021_2617_MOESM2_ESM.tif
Supplementary file2 The maximum-likelihood tree of deduced amino acid sequence of PmMCA. The phylogenetic position of present P. minimum is marked with a red color. The amino acid sequences used in this study were obtained from GenBank database. (TIF 893 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Ki, JS. Identification of a Metacaspase Gene in the Bloom-Forming Dinoflagellate Prorocentrum minimum and its Putative Function Involved in Programmed Cell Death. Curr Microbiol 78, 3577–3585 (2021). https://doi.org/10.1007/s00284-021-02617-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02617-3