Abstract
Background and aims
Gut microbiota and their metabolic products might play important roles in regulating the pathogenesis of choledocholithiasis concurrent with cholangitis (CC). The aim of this study was to explore the characteristic gut dysbiosis, metabolite profiles and the possible roles in patients with CC.
Methods
A case–control study was carried out to analyze the alterations in the intestinal microbiota and their metabolites in patients with CC (n = 25) compared with healthy controls (HCs) (n = 25) by metagenomic sequencing to define the gut microbiota community and liquid chromatography/mass spectrometry (LC/MS) analysis to characterize the metabolite profiles.
Results
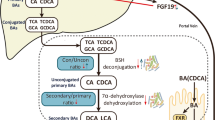
Significantly reduced Shannon diversity index (p = 0.043) and differential overall fecal microbiota community in CCs were observed. Twelve dominant altered species were identified and analyzed (LDA score > 3.0, p < 0.05) (Q value < 0.05), including unclassified_f_Enterobacteriaceae, Escherichia_coli, Roseburia_faecis and Eubacterium rectale. Moreover, the levels of KEGG pathways related to biofilm formation of Escherichia coli, lipopolysaccharide (LPS) biosynthesis, and the metabolism of propanoate and glutathione in CCs were significantly altered. Finally, 47 markedly changed metabolites (VIP > 1.0 and p < 0.05), including low level of kynurenic acid (KYNA) and high concentration of N-palmitoylsphingosine involving tryptophan metabolism and sphingolipid signaling pathways, were identified to validate aberrant metabolic patterns in CCs, and multiple correlated metabolic modules involving bile inflammation were altered in CCs.
Conclusion
Our study provides novel insights into compositional and functional alterations in the gut microbiome and metabolite profiles in CC and the underlying mechanisms between gut microbiota and bile inflammation.






Similar content being viewed by others
Data availability
The metagenome sequencing data are available (at https://cloud.majorbio.com). The data from Changzheng Hospital are available upon reasonable request from the corresponding author (Y.L.). The data from Changzheng Hospital are not publicly available, because they contain protected patient privacy information. The other data relevant to the study are included in the article or uploaded as supplementary information.
Abbreviations
- CC:
-
Choledocholithiasis concurrent with cholangitis
- HC:
-
Healthy control
- ERCP:
-
Endoscopic retrograde cholangiopancreatography
- SCFA:
-
Short-chain fatty acid
- NAFLD:
-
Nonalcoholic fatty liver disease
- LC/MS:
-
Chromatography/mass spectrometry
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- QIIME:
-
Quantitative insights into microbial ecology
- PCoA:
-
Principal coordinate analysis
- LDA:
-
Linear discriminant analysis
- LEfSe:
-
Linear discriminant analysis effect size
- VIP:
-
Variable importance in the projection
- WBC:
-
White blood cell
- CRP:
-
C-reaction protein
- KYNA:
-
Kynurenic acid
- TLRs:
-
Toll-like receptors
- NO:
-
Nitric oxide
- LPS:
-
Lipopolysaccharide
References
Frossard JL, Morel PM. Detection and management of bile duct stones. Gastrointest Endosc. 2010;72(4):808–16. https://doi.org/10.1016/j.gie.2010.06.033.
Peery AF, Crockett SD, Barritt AS, et al. Burden of gastrointestinal, liver, and pancreatic diseases in the United States. Gastroenterology. 2015;149(7):1731-1741.e3. https://doi.org/10.1053/j.gastro.2015.08.045.
ASGE Standards of Practice Committee, Buxbaum JL, Abbas Fehmi SM, et al. ASGE guideline on the role of endoscopy in the evaluation and management of choledocholithiasis. Gastrointest Endosc. 2019;89(6):1075–105. https://doi.org/10.1016/j.gie.2018.10.001.
Lai EC, Mok FP, Tan ES, et al. Endoscopic biliary drainage for severe acute cholangitis. N Engl J Med. 1992;326(24):1582–6. https://doi.org/10.1056/NEJM199206113262401.
Negm AA, Schott A, Vonberg RP, et al. Routine bile collection for microbiological analysis during cholangiography and its impact on the management of cholangitis. Gastrointest Endosc. 2010;72(2):284–91. https://doi.org/10.1016/j.gie.2010.02.043.
Lammert F, Gurusamy K, Ko CW, et al. Gallstones. Nat Rev Dis Primers. 2016;2:16024. https://doi.org/10.1038/nrdp.2016.24.
Van Erpecum KJ, Van Berge-Henegouwen GP. Gallstones: an intestinal disease? Gut. 1999;44(3):435–8. https://doi.org/10.1136/gut.44.3.435.
Wang HH, Portincasa P, Mendez-Sanchez N, et al. Effect of ezetimibe on the prevention and dissolution of cholesterol gallstones. Gastroenterology. 2008;134(7):2101–10. https://doi.org/10.1053/j.gastro.2008.03.011.
Vítek L, Carey MC. New pathophysiological concepts underlying pathogenesis of pigment gallstones. Clin Res Hepatol Gastroenterol. 2012;36(2):122–9. https://doi.org/10.1016/j.clinre.2011.08.010.
Tripathi A, Debelius J, Brenner DA, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15(7):397–411. https://doi.org/10.1038/s41575-018-0011-z.
Sun J, Chang EB. Exploring gut microbes in human health and disease: pushing the envelope. Genes Dis. 2014;1(2):132–9. https://doi.org/10.1016/j.gendis.2014.08.001.
Wang Q, Jiao L, He C, et al. Alteration of gut microbiota in association with cholesterol gallstone formation in mice. BMC Gastroenterol. 2017;17(1):74. https://doi.org/10.1186/s12876-017-0629-2.
Verdier J, Luedde T, Sellge G. Biliary mucosal barrier and microbiome. Viszeralmedizin. 2015;31(3):156–61. https://doi.org/10.1159/000431071.
Sayin SI, Wahlström A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–35. https://doi.org/10.1016/j.cmet.2013.01.003.
Wu T, Zhang Z, Liu B, et al. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics. 2013;14:669. https://doi.org/10.1186/1471-2164-14-669.
Liu J, Yan Q, Luo F, et al. Acute cholecystitis associated with infection of Enterobacteriaceae from gut microbiota. Clin Microbiol Infect. 2015;21(9):851.e1-9. https://doi.org/10.1016/j.cmi.2015.05.017.
Capoor MR, Nair D, Rajni, et al. Microflora of bile aspirates in patients with acute cholecystitis with or without cholelithiasis: a tropical experience. Braz J Infect Dis. 2008;12(3):222–5. https://doi.org/10.1590/s1413-86702008000300012.
Chen B, Fu SW, Lu L, et al. A preliminary study of biliary microbiota in patients with bile duct stones or distal cholangiocarcinoma. Biomed Res Int. 2019;2019:1092563. https://doi.org/10.1155/2019/1092563.
Feng J, Zhao F, Sun J, et al. Alterations in the gut microbiota and metabolite profiles of thyroid carcinoma patients. Int J Cancer. 2019;144(11):2728–45. https://doi.org/10.1002/ijc.32007.
Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72(3):558–77. https://doi.org/10.1016/j.jhep.2019.10.003.
Wei Y, Li Y, Yan L, et al. Alterations of gut microbiome in autoimmune hepatitis. Gut. 2020;69(3):569–77. https://doi.org/10.1136/gutjnl-2018-317836.
Xue F, Nan X, Sun F, et al. Metagenome sequencing to analyze the impacts of thiamine supplementation on ruminal fungi in dairy cows fed high-concentrate diets. AMB Express. 2018;8(1):159. https://doi.org/10.1186/s13568-018-0680-6.
Li R, Li Y, Kristiansen K, et al. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–4. https://doi.org/10.1093/bioinformatics/btn025.
Peng W, Yi P, Yang J, et al. Association of gut microbiota composition and function with a senescence-accelerated mouse model of Alzheimer’s Disease using 16S rRNA gene and metagenomic sequencing analysis. Aging (Albany NY). 2018;10(12):4054–65. https://doi.org/10.18632/aging.101693.
Quinn RA, Melnik AV, Vrbanac A, et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature. 2020;579(7797):123–9. https://doi.org/10.1038/s41586-020-2047-9.
Adams LA, Wang Z, Liddle C, et al. Bile acids associate with specific gut microbiota, low-level alcohol consumption and liver fibrosis in patients with non-alcoholic fatty liver disease. Liver Int. 2020;40(6):1356–65. https://doi.org/10.1111/liv.14453.
Cresci GAM. Is it time to consider gut microbiome readouts for precision diagnosis and treatment of alcoholic liver disease? Hepatology. 2020;72(1):4–6. https://doi.org/10.1002/hep.31245.
Schwabe RF, Greten TF. Gut microbiome in HCC—mechanisms, diagnosis and therapy. J Hepatol. 2020;72(2):230–8. https://doi.org/10.1016/j.jhep.2019.08.016.
Friedfeld MR, Zhong H, Ruck RT, et al. Cobalt-catalyzed asymmetric hydrogenation of enamides enabled by single-electron reduction. Science. 2018;360(6391):888–93. https://doi.org/10.1126/science.aar6117.
Ren Z, Li A, Jiang J, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68(6):1014–23. https://doi.org/10.1136/gutjnl-2017-315084.
Di Ciaula A, Wang DQ, Portincasa P. An update on the pathogenesis of cholesterol gallstone disease. Curr Opin Gastroenterol. 2018;34(2):71–80. https://doi.org/10.1097/MOG.0000000000000423.
Levy M, Kolodziejczyk AA, Thaiss CA, et al. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17(4):219–32. https://doi.org/10.1038/nri.2017.7.
Razaghi M, Tajeddin E, Ganji L, et al. Colonization, resistance to bile, and virulence properties of Escherichia coli strains: unusual characteristics associated with biliary tract diseases. Microb Pathog. 2017;111:262–8. https://doi.org/10.1016/j.micpath.2017.08.043.
Zhao J, Wang Q, Zhang J. Changes in microbial profiles and antibiotic resistance patterns in patients with biliary tract infection over a six-year period. Surg Infect (Larchmt). 2019;20(6):480–5. https://doi.org/10.1089/sur.2019.041.
Mizutani T, Mitsuoka T. Inhibitory effect of some intestinal bacteria on liver tumorigenesis in gnotobiotic C3H/He male mice. Cancer Lett. 1980;11(2):89–95. https://doi.org/10.1016/0304-3835(80)90098-1.
Pittayanon R, Lau JT, Leontiadis GI, et al. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology. 2020;158(4):930-946.e1. https://doi.org/10.1053/j.gastro.2019.11.294.
Li KY, Wang JL, Wei JP, et al. Fecal microbiota in pouchitis and ulcerative colitis. World J Gastroenterol. 2016;22(40):8929–39. https://doi.org/10.3748/wjg.v22.i40.8929.
David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. https://doi.org/10.1038/nature12820.
Chia JH, Feng Y, Su LH, et al. Clostridium innocuum is a significant vancomycin-resistant pathogen for extraintestinal clostridial infection. Clin Microbiol Infect. 2017;23(8):560–6. https://doi.org/10.1016/j.cmi.2017.02.025.
Feng Q, Liu Z, Zhong S, et al. Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease. Sci Rep. 2016;6:22525. https://doi.org/10.1038/srep22525.
Ferrero MA, Martínez-Blanco H, Lopez-Velasco FF, et al. Purification and characterization of GlcNAc-6-P 2-epimerase from Escherichia coli K92. Acta Biochim Pol. 2007;54(2):387–99.
Martin G, Kolida S, Marchesi JR, et al. In vitro modeling of bile acid processing by the human fecal microbiota. Front Microbiol. 2018;9:1153. https://doi.org/10.3389/fmicb.2018.01153.
Lee JH, Regmi SC, Kim JA, et al. Apple flavonoid phloretin inhibits Escherichia coli O157:H7 biofilm formation and ameliorates colon inflammation in rats. Infect Immun. 2011;79(12):4819–4127. https://doi.org/10.1128/IAI.05580-11.
Niu GC, Liu L, Zheng L, et al. Mesenchymal stem cell transplantation improves chronic colitis-associated complications through inhibiting the activity of toll-like receptor-4 in mice. BMC Gastroenterol. 2018;18(1):127. https://doi.org/10.1186/s12876-018-0850-7.
Yokoyama T, Komori A, Nakamura M, et al. Human intrahepatic biliary epithelial cells function in innate immunity by producing IL-6 and IL-8 via the TLR4-NF-kappaB and -MAPK signaling pathways. Liver Int. 2006;26(4):467–76. https://doi.org/10.1111/j.1478-3231.2006.01254.x.
Zhang Y, Yu W, Han D, et al. L-lysine ameliorates sepsis-induced acute lung injury in a lipopolysaccharide-induced mouse model. Biomed Pharmacother. 2019;118: 109307. https://doi.org/10.1016/j.biopha.2019.109307.
Jiao N, Baker SS, Nugent CA, et al. Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: a meta-analysis. Physiol Genomics. 2018;50(4):244–54. https://doi.org/10.1152/physiolgenomics.00114.2017.
Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–5. https://doi.org/10.1038/nature12726.
Cani PD. Is colonic propionate delivery a novel solution to improve metabolism and inflammation in overweight or obese subjects? Gut. 2019;68(8):1352–3. https://doi.org/10.1136/gutjnl-2019-318776.
Ciocan D, Voican CS, Wrzosek L, et al. Bile acid homeostasis and intestinal dysbiosis in alcoholic hepatitis. Aliment Pharmacol Ther. 2018;48(9):961–74. https://doi.org/10.1111/apt.14949.
Wirthgen E, Hoeflich A, Rebl A, et al. Kynurenic Acid: the Janus-faced role of an immunomodulatory tryptophan metabolite and its link to pathological conditions. Front Immunol. 2018;8:1957. https://doi.org/10.3389/fimmu.2017.01957.
Marciniak S, Wnorowski A, Smolińska K, et al. Kynurenic Acid protects against thioacetamide-induced liver injury in rats. Anal Cell Pathol (Amst). 2018;2018:1270483. https://doi.org/10.1155/2018/1270483.
Moroni F, Cozzi A, Sili M, et al. Kynurenic acid: a metabolite with multiple actions and multiple targets in brain and periphery. J Neural Transm (Vienna). 2012;119(2):133–9. https://doi.org/10.1007/s00702-011-0763-x.
Gaffen SL, Jain R, Garg AV, et al. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14(9):585–600. https://doi.org/10.1038/nri3707.
Raichur S, Brunner B, Bielohuby M, et al. The role of C16:0 ceramide in the development of obesity and type 2 diabetes: CerS6 inhibition as a novel therapeutic approach. Mol Metab. 2019;21:36–50. https://doi.org/10.1016/j.molmet.2018.12.008.
Albeituni S, Stiban J. Roles of ceramides and other sphingolipids in immune cell function and inflammation. Adv Exp Med Biol. 2019;1161:169–91. https://doi.org/10.1007/978-3-030-21735-8_15.
Marra F, Svegliati-Baroni G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J Hepatol. 2018;68(2):280–95. https://doi.org/10.1016/j.jhep.2017.11.014.
Chaurasia B, Tippetts TS, Mayoral Monibas R, et al. Targeting a ceramide double bond improves insulin resistance and hepatic steatosis. Science. 2019;365(6451):386–92. https://doi.org/10.1126/science.aav3722.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81670544, 81870416), the Top-Level Clinical Discipline Project of Shanghai Pudong (PWYgf2018-04), the Science and Technology Guidance Plan of Shanghai Science and Technology Commission (19411970500) and the Pilot Talent Plan of Shanghai East Hospital (2019lhrcjh).
Author information
Authors and Affiliations
Contributions
Designed the experiments: XZ, and YL. Collected the clinical samples: KGT, KMW, and PMS. Performed the microbiome analysis: ZYH, YYL, and YTL. Performed the metabolomic analysis: BBL, and PQW. Draft the manuscript: ZYH, and YL. Revised the manuscript for intellectual content: ZYH, KGT, PQW, XZ and YL.
Corresponding authors
Ethics declarations
Conflict of interest
No potential conflicts of interest were disclosed by Zhiyuan Hao, Kegong Tao, Kaiming Wu, Yuanyuan Luo, Yiting Lu, Binbin Li, Peimei Shi, Peiqin Wang, Xin Zeng, Yong Lin.
Ethics approval
The study was approved by the Institutional Ethics Committee of Shanghai Changzheng Hospital. Informed consent was obtained prior to data collection.
Patient and public involvement
Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication
Not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hao, Z., Tao, K., Wu, K. et al. Alterations of gut microbiome and metabolite profiles in choledocholithiasis concurrent with cholangitis. Hepatol Int 16, 447–462 (2022). https://doi.org/10.1007/s12072-021-10231-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-021-10231-5