Abstract
Male breast cancer (BCa) is a rare disease accounting for less than 1% of all breast cancers and 1% of all cancers in males. The clinical management is largely extrapolated from female BCa. Several multigene assays are increasingly used to guide clinical treatment decisions in female BCa, however, there are limited data on the utility of these tests in male BCa. Here we present the gene expression results of 381 M0, ER+ve, HER2-ve male BCa patients enrolled in the Part 1 (retrospective analysis) of the International Male Breast Cancer Program. Using a custom NanoString™ panel comprised of the genes from the commercial risk tests Prosigna®, OncotypeDX®, and MammaPrint®, risk scores and intrinsic subtyping data were generated to recapitulate the commercial tests as described by us previously. We also examined the prognostic value of other risk scores such as the Genomic Grade Index (GGI), IHC4-mRNA and our prognostic 95-gene signature. In this sample set of male BCa, we demonstrated prognostic utility on univariate analysis. Across all signatures, patients whose samples were identified as low-risk experienced better outcomes than intermediate-risk, with those classed as high risk experiencing the poorest outcomes. As seen with female BCa, the concordance between tests was poor, with C-index values ranging from 40.3% to 78.2% and Kappa values ranging from 0.17 to 0.58. To our knowledge, this is the largest study of male breast cancers assayed to generate risk scores of the current commercial and academic risk tests demonstrating comparable clinical utility to female BCa.
Similar content being viewed by others
Introduction
Male breast cancer (BCa) represents ~1% of all newly diagnosed cancers in men1 and ~1% of all breast cancers2. Research into this rare disease has been limited, with treatment largely extrapolated from knowledge about female BCa3. Surgical management is usually modified radical mastectomy, with a minority of patients being offered breast conserving treatment4. Local and systemic treatment is largely informed by treatment indications and regimens used in female breast cancer. However, for adjuvant endocrine treatment, the use of aromatase inhibitors (AIs) alone is not recommended, with tamoxifen for at least 5 years indicated for ER/PgR positive tumors3,5. Where AIs are indicated, for example in metastatic male BCa, pituitary blockade with an LHRH agonist or orchiectomy is recommended6.
Genetic counselling is recommended for all men with BCa, regardless of family history, due to strong links between male BCa and BRCA2 mutations, seen in 10% of men with BCa3,7,8,9. Transcriptomic multiparametric assays are now integrated into clinical management guidelines for early female BCa10 both as prognostic tools and to identify patients for adjuvant chemotherapy11,12. Most guidelines refer exclusively to female BCas with respect to the use of these multiparametric assays. Data relating to these tests in male BC are from retrospective series, most with small numbers of cases limited to evaluation of prognosis for single tests13,14,15,16; however, we are not aware of any analyses which provide comparative data on multiple signatures with respect to patient outcome. We developed a method to compare signatures using a combined quantitative mRNA array covering key molecular signatures17, which have been trained against the results of the same signatures measured by the original methodology18. We describe here an analysis of male BCa samples from the EORTC cohort19 using these “trained” signatures to compare the result of each test and to determine the association between test result and prognosis in the context of a multi-institutional male BCa cohort.
Results
mRNA profiling
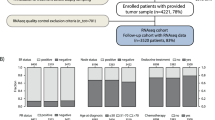
Out of the 1483 patients included in the parental study, 699 (47.1%) patients met the eligibility criteria of the research project; the main reasons for exclusion were missing tissue, event status, or event dates (see Supplementary Fig. 1). As previously reported19, no evidence for a selection bias due to missing data has been identified. From these, 389 samples had sufficient material for extraction and 381 samples yielded sufficient RNA.
All 381 samples assayed were successfully analyzed using the custom NanoString gene expression panel and passed the quality control (Supplementary Fig. 1, Supplementary Table 1).
Distribution of gene signatures and concordance
Table 1 details the distribution of risk classification across tests, which markedly differs from one gene signature to another. The proportion of high risk patients ranged from 15.7 to 63.5% and for low risk patients the range was 9.4–53.5% (Table 1). For tests with 3 risk groups, the proportion of intermediate risk cases ranged from 38.3 to 55.6%. Low risk cases ranged from 9.4 to 53.5% of all cases and high risk cases from 15.7 to 63.5%. Cross-tabulations for all test combinations are shown in Supplementary Tables 2–15. When cross-tabulating the pairs of gene signatures, the C-index values ranged from 40.3 to 78.2% and kappa values ranged from 0.17 to 0.58, indicating slight to moderate agreement between the gene signatures (Table 2). Using the Prosigna-trained sub-type classification, 0.8% of cases were Basal-like, 3.4% HER2-enriched, 20.2% Luminal A and 75.6% Luminal B.
Survival analysis
Seventy-four patients experienced a locoregional recurrence or distant progression qualifying as events for TTR endpoint, of whom, 55 (74.3%) reported a distant progression as first event. Sixty-one patients experienced a distant recurrence qualifying as events for the TTDR endpoint and 38 patients died after a distant recurrence (BCSS endpoint). Seventy-four patients died in the absence of a distant progression and these deaths were considered as competing risks.
Also critical to this analysis, with respect to outcome, when competing risks of deaths not preceded by distant recurrence were accounted for, the cumulative incidence of specific BCa-related events were consistently lower in patients classified as low risk. The 5-year cumulative incidence of locoregional or distant recurrence in low risk patients ranged from 3.7% (95%CI: 0.3–16.3) (Prosigna-trained) to 10.2% (95%CI: 5.6–16.2) (Risk 95-Gene; Figs. 1–2, Supplementary Figs. 2–5) and from 3.7% (95%CI: 0.3–16.3) (Prosigna-trained) to 8.2% (95%CI: 4.1–14.0) (Risk 95-gene) for distant recurrence. For high risk patients, 5-year cumulative incidence of locoregional or distant recurrences ranged from 24.6% (95%CI: 13.8–37.1) (IHC4-RNA) to 20.0% (95%CI: 14.2–26.5) (95-gene) and from 22.1% (95%CI: 15.4–29.7) (Prosigna-trained) to 17.8% (95%CI: 12.2–24.2) (95-gene) for distant recurrence. Regarding BCSS, the 5-year cumulative incidence was below 5% for low risk patients for all tests, whilst for high risk patients the rates ranged between 9.8% (95%CI: 5.8–14.9) and 13.7% (95%CI: 7.1–22.4).
a Oncotype DX-trained risk classification for Time to Relapse; b Oncotype DX-trained risk classification for Time to Distant Relapse; c Oncotype DX-trained risk classification for Breast Cancer Specific Survival. Cumulative incidence rates for low (blue line), intermediate (green line), and high (red line) Oncotype DX-trained results with corresponding 95%CI (shaded areas) estimated by cumulative incidence function accounting for deaths not preceded by a distant relapse as competing risks. Total events/risk group (Events/Total) represent all events observed during follow up (up to 12 years). CIF = 5-year cumulative event frequency (percent) at 5 years with estimated 95% confidence intervals (95% CI).
a ROR-PT-trained risk classification for Time to Relapse; b ROR-PT-trained risk classification for Time to Distant Relapse; c ROR-PT-trained risk classification for Breast Cancer Specific Survival. Cumulative incidence rates for low (blue line), intermediate (green line), and high (red line) ROR-PT-trained results with corresponding 95%CI (shaded areas) estimated by cumulative incidence function accounting for deaths not preceded by a distant relapse as competing risks. Total events/risk group (Events/Total) represent all events observed during follow up (up to 12 years). CIF = 5-year cumulative event frequency (percent) at 5 years with estimated 95% confidence intervals (95% CI).
For all signatures evaluated, there was evidence of prognostic information associated with statistically significant risk stratification in low, intermediate (where applicable), and high risk groups (Figs. 1–2, Supplementary Figs. 2–5, Table 3). Each signature provided statistically significant separation of patients into low, intermediate (where appropriate) and high risk groups with respect to TTR, TTDR, and BCSS in univariate analyses (Gray test < 0.01, except for the IHC RNA4 signature) (Figs. 1–2, Supplementary Figs. 2–5). On the contrary, in multivariate-adjusted analyses for TTR and TTDR, the effect of gene-signatures was no longer significant (Table 3).
The 5-year AUC from time-dependent ROC analysis ranged from 0.63 (95%CI: 0.54–0.72) (IHC4-RNA) to 0.72 (95%CI: 0.64–0.80) (Prosigna-trained) for TTR and from 0.65 (95%CI: 0.55–0.74) (IHC4-RNA) to 0.73 (95%CI: 0.64–0.82) (Prosigna-trained) for TTDR. Regarding BCSS, AUC is about 0.75 for each gene signature and is maximal for Risk-95 gene signature (AUC = 0.82, 95%CI: 0.71–0.93) (Supplementary Figs. 6–8).
Discussion
The use of molecular prognostic assays in female BCa is now well established, but evidence relating to their performance in male BCa patients is sparse. In this study we show, using computational methods to recapitulate multiple BCa prognostic signatures, evidence for the prognostic impact of multiple gene signatures. However, we also show evidence of discordance between signatures, applied to the same case, similar to that seen in female BCa20. This study highlights the potential utility of molecular prognostic signatures in male breast cancer but suggests that more research is needed if we are to fully understand the potential value of different approaches to assessing prognosis and directing treatment, using molecular tools, in men with breast cancer.
Critical to our study is the close correlation between the computationally derived “signature trained” scores and true results as shown in our recent paper18. For ROR-PT results the correlation coefficient between “trained” and true assay results was 0.93, comparing true to “trained” results showed 90% of cases within the same risk category (low, intermediate, high—see ref. 18). Similarly for “Oncotype Dx-trained” results the correlation coefficient between true and “trained” results was 0.87 with 75% of results giving the same risk category (see ref. 18) and only 1% of cases disagreeing by more than 1 risk category. For MammaPrint -trained results, which were calculated only as categorical high versus low risk groups, over 90% of cases were classified in the same risk group by “trained” and true results18. Full details of these results are reported elsewhere18. For Genomic Grade Index and IHC4, we did not have access to actual assay results to enable us to train signatures as we did for other signatures, and for Endopredict we did not have enough genes covered to allow recapitulation of this signature.
Using a common analysis platform and computational methods to recapitulate prognostic scores we found that all molecular signatures tested: Oncotype DX-trained21,22, Prosigna-ROR-PT23,24, MammaPrint25,26,27, Genomic Grade Index28, IHC4-mRNA based IHC429, and our novel 95-gene signature17 demonstrated the ability to segregate male BCas into high and low prognostic risk groupings. All signatures were associated with significant differences in 5-year survival for time to recurrence, time to distant relapse and breast cancer-specific survival, between low and high risk groups in univariate analyses (Figs. 1–2, Supplementary Figs. 2–5). However, due to the relatively small number of breast cancer-specific events, we were unable to demonstrate the statistically significant prognostic impact of the majority of signatures in multivariate analysis when adjusting for the following key clinico-pathological covariates:age, grade, nodal status and tumor size and treatment variables (adjuvant chemotherapy, radiotherapy, endocrine treatment). There have been few reports on the utility of prognostic signatures, developed using female BCas, when applied to male BCas and these have largely focused on the utility of Oncotype DX13,15. Previous studies showed an 81% 5-year BCa-specific survival for men with recurrence scores >31, slightly lower than the 86.3% 5-year BCSS observed for men with recurrence scores >25 shown in the current study, but commensurate with the different thresholds used13,15. In the study by Massarweh et al.15, 27.8% of men exhibited RS > 25 compared with 12.4% with scores >31. Given the modest number of events in both studies, we believe our results are broadly comparable to those reported by Massarweh et al.15. Results from a similar study by Wang et al.14 show a higher all-cause mortality rate in all risk groups, but is limited by failure to exclude competing causes of death, which accounted for almost 50% of events in our current study. This high percentage may be due to the fact that male BC patients are older and have more co-morbidities than their female counterparts. We are unaware of studies reporting patient outcome in male BCa when stratified by tests other than Oncotype DX, making it more challenging to draw comparisons between studies using these molecular assays. With respect to the 50-gene signature driving molecular subtypes (Prosigna/PAM50), a study by Sanchez-Munoz et al.16, profiled 67 invasive male BCas using the NanoString panel identifying 60% of cases as Luminal B, 30% Luminal A and 10% HER2-enriched; which is consistent with our findings18, however, we are not aware of any studies of male BCas profiled reporting Prosigna risk scores.
As with prior comparisons in female BCa20, we demonstrate poor agreement between risk signatures in male BCa with kappa values ranging from 0.17 to 0.58 (Table 2). This modest agreement reinforces observations from larger cohorts of female BCas that different molecular risk scores based on limited mRNA panels may not capture all features related to risk in this population. This conclusion is supported by multiple analyses showing the added value of combining multiple risk signatures in female BCa30 and our own recent data highlighting the modest AUCs associated with different molecular signatures17 with respect to predicting outcome. Despite different methodologies used, AUCs of time-dependent ROC curves at 5 years for male breast cancer cases fall within the same range of the AUCs reported in female patients17. This provides no indication that different cut points for risk would apply to male rather than female breast cancer, however, given the small sample size of the present study it is premature to exclude this possibility entirely. All signatures assessed would appear appropriate for use in male breast cancer patients.
There are several key limitations to our current research project. Firstly, we have used computational methods to calculate the relevant risk signatures rather than the original assays as used in the clinical setting. This limitation is offset in part by the use of a training and validation approach to benchmark results for Oncotype DX, Prosigna, and MammaPrint results against true assay results18, but remains a limitation for other tests. Secondly, the analyses were conducted in a retrospective dataset in which not all data were systematically collected in all patients including, the cause of death is not reported for a substantial number of patients leading to a substantial proportion of competing risks. As a result, we have not presented overall survival (all causes) data since this would be confounded by the lack of data as to cause of death in many patients. Despite this study representing one of the largest cohorts of male breast cancers analyzed to date, the sample size and in particular the number of breast cancer related events, limit the statistical power of this analysis. In particular, we were not able to compare the impact of multiple tests performed in sequence due to a lack of statistical power nor were we able to assess the potential impact of tests on chemoprediction. Notwithstanding these limitations we are able to show the ability of a number of existing multiparametric tests (including MammaPrint, Oncotype Dx, Prosigna ROR-PT, Genomic Grade Index and a novel 95-gene signature) to provide useful prognostic information in male breast cancer. These data provide evidence to support the utility of multiple prognostic assays in the context of male breast cancer. Further research to identify the optimal prognostic approach to male breast cancer, perhaps including genomic features such as mutations and copy number alterations, is warranted in addition to investigating the role of intratumoural heterogeneity.
Methods
Patients and samples
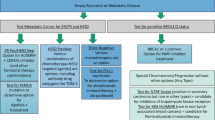
The retrospective cohort study of the EORTC/TBCRC/BIG/NABCG International Male Breast Cancer Program enrolled male patients with histologically proven BCa, diagnosed between 1990 and 2010, across multiple participating institutions19. Ethics approval was provided by the University of Toronto (#30035), a waiver of consent was approved since patient contact was not feasible due to death or loss to follow-up and the research involved no risk to patients whose identify was coded and confidentially protected. Patients with all disease stages (early, locally advanced, and metastatic) were included, irrespective of the treatment received. Availability of a tissue sample (Formalin-Fixed-Paraffin-Embedded—FFPE) of good quality was mandatory for enrollment. Biological material was handled and analyzed centrally according to published guidelines for adoption across BCa clinical trials, conducted by BIG and NABCG, in 200831. Patients in this research project were selected from the retrospective cohort study based on the following exclusion criteria: patients ineligible for the analysis of the parental retrospective cohort, with metastatic (M1/MX) disease, ER-ve per central pathology or local pathology (if central pathology not available), HER2+ve or unknown based on central pathology, insufficient information for assessment of recurrence free survival. In addition, samples with insufficient RNA or which failed the quality control criteria were excluded. All institutions participating in the retrospective cohort study obtained ethical approval from their institutions including consent waivers.
RNA extraction and expression profiling using NanoString
Profiling of all samples was performed using mRNA extracted and analyzed using the NanoString codeset as described previously17 at the Ontario Institute for Cancer Research (OICR).
Derivation of signature-trained risk stratification scores from candidate assays
Based on our study comparing two different approaches to the generation of simulated risk scores18 we selected a training and validation approach based on results obtained from the OPTIMA prelim study20 to best fit risk stratification scores generated for this study to those derived from the relevant commercial assay. For all tests we used the suffix “-trained” to discriminate the computationally derived assays scores from the commercially derived scores, e.g., Oncotype DX-trained vs. Oncotype-DX®. For each of the commercial test, cases were grouped into pre-defined risk categories according to the cut-points: Oncotype DX—low risk <18, intermediate risk 18–25, high risk ≥25; Prosigna—low risk <40, intermediate risk 40–60, high risk ≥61; MammaPrint—low risk and high risk as described in ref. 18. We modified the original cut point for “high risk” for the Oncotype DX test in line with reported results from the TAILORx trial11,32 and our previous reported results from OPTIMA prelim20. For “Prosigna”, results refer throughout to the ROR-PT risk score in clinical use, which includes tumor pathological size. For the Genomic Grade Index (GGI), the suffix “-like” refers to recapitulation of the risk score as previously described though not trained against a benchmark dataset. The IHC4-mRNA signature is similarly modelled to estimate risk by the transcriptomic expression of ER, PgR, Ki67, and HER2 originally based on the immunohistochemical signature described by Cuzick et al.33 The 95-gene signature has been previously described by our group18.
Statistical analyses
Results from the expression profiling using NanoString were provided to EORTC to perform the statistical analysis of clinical data, long term outcomes, and local and central pathology data. Descriptive statistical analysis was performed for patient characteristics, disease characteristics, and treatment(s) administered.
Cross-tabulation of risk classification (low, intermediate—where applicable, high) as defined by the different gene signatures were tabulated to assess concordance and agreement of classification across the different gene signatures. Concordance index and kappa agreement coefficients and their corresponding 95% confidence interval (CI) were estimated. When cross-tabulating gene signatures with different number of categories (i.e., three categories such as low, intermediate, high versus two categories such as high, low), the intermediate category was combined with the low category and Cohen’s simple kappa was estimated while for ternary versus ternary comparisons, the weighted kappa was used.
The prognostic value of the gene signatures was assessed for the following endpoints: time to distant relapse (TTDR) defined as the time until the first distant progression, time to relapse (TTR) defined as the time until the first loco-regional recurrence or distant progression, breast cancer-specific survival (BCSS) defined as the time until breast cancer related death, considering death preceded by a distant relapse. For these endpoints, deaths in the absence of distant relapse are considered as competing risk. The endpoints were calculated from the time of first diagnosis of BCa. Patients without an event for the above endpoints were censored at the last date known alive.
The event rates at 5 years and corresponding 95% confidence intervals were estimated by the cumulative incidence method. Cumulative incidence functions between the risk groups were compared based on the Gray test at a significance level of 0.05. Fine and Gray models were used to estimate the univariate and adjusted hazard ratio (HR) and their corresponding 95%CI. The multivariate models were adjusted for known prognostic clinico-pathological variables (age, grade, nodal status and tumor size) and treatment variables (adjuvant chemotherapy, radiotherapy, endocrine treatment) and the multivariate p-value was estimated with the use of a Wald test. Due to the low number of events for BCSS, only univariate analyses were conducted for this endpoint. The proportional hazard assumption was checked graphically using a plot of the log cumulative hazard. The analyses were not adjusted for multiple testing.
The ability of the gene signatures to predict clinical outcome at 5 years was assessed by time-dependent receiver operating characteristic curves (ROC) and the corresponding area under the curve (AUC).The underlying method of ROC curves has been extended to the setting of censored observations and presence of competing risks34. Time-dependent ROC curves at 5 years were plotted and the corresponding AUCs estimated for each endpoint (Time to relapse, Time to distant relapse, Breast Cancer-specific survival) and for each gene signature to the exception of MammaPrint. As described previously18 when training the algorithm for MammaPrint, only dichotomized risk categories were available preventing any AUC analysis with this signature. Cases were patients that experienced the event of interest in the first five years of follow-up, while controls were defined as patients that were either event-free at 5 years, or experienced a competing event in the first 5 years of follow-up.
Analyses were performed with SAS software, version 9.4 (SAS Institute) and the time-dependent ROC curves were plotted in R, version 4.0.0, with the timeROC package.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data generated and analyzed during this study are described in the following data record: https://doi.org/10.6084/m9.figshare.1461683135. The data underlying the Kaplan–Meier survival curves and tables that support the findings of this study are available from European Organisation for the Research and Treatment of Cancer (EORTC). However, the data are not publicly available and restrictions apply to their availability as they were used under license from EORTC for the current study. Data can be made available with the permission of EORTC. Data enquiries can be made to the corresponding author, and data requests can be made at https://www.eortc.org/data-sharing/.
Code availability
The codes that support these findings are subject to patent applications and restrictions related to licenses. Codes are available from the authors upon reasonable request and with the permission of the Ontario Institute for Cancer Research (OICR).
References
Anderson, W. F., Jatoi, I., Tse, J. & Rosenberg, P. S. Male breast cancer: a population-based comparison with female breast cancer. J. Clin. Oncol. 28, 232–239 (2010).
Giordano, S. H., Cohen, D. S., Buzdar, A. U., Perkins, G. & Hortobagyi, G. N. Breast carcinoma in men: a population-based study. Cancer 101, 51–57 (2004).
Korde, L. A. et al. Multidisciplinary meeting on male breast cancer: summary and research recommendations. J. Clin. Oncol. 28, 2114–2122 (2010).
Cutuli, B. Strategies in treating male breast cancer. Expert. Opin. Pharmacother. 8, 193–202 (2007).
Hassett, M. J. et al. Management of male breast cancer: ASCO Guideline. J Clin Oncol 38, 1849–1863 (2020).
Cardoso, F. et al. ESO-ESMO 2nd international consensus guidelines for advanced breast cancer (ABC2)dagger. Ann. Oncol. 25, 1871–1888 (2014).
Ewertz, M., Holmberg, L., Tretli, S., Pedersen, B. V. & Kristensen, A. Risk factors for male breast cancer–a case-control study from Scandinavia. Acta Oncol. 40, 467–471 (2001).
Liede, A., Karlan, B. Y. & Narod, S. A. Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: a review of the literature. J. Clin. Oncol. 22, 735–742 (2004).
Meijers-Heijboer, H. et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat. Genet. 31, 55–59 (2002).
Vieira, A. F. & Schmitt, F. An update on breast cancer multigene prognostic tests—emergent clinical biomarkers. Fronti. Med. https://doi.org/10.3389/fmed.2018.00248 (2018).
Sparano, J. A. et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N. Eng. J. Med. 379, 111–121 (2018).
Sestak, I. et al. Prediction of chemotherapy benefit by EndoPredict in patients with breast cancer who received adjuvant endocrine therapy plus chemotherapy or endocrine therapy alone. Breast Cancer Res. Treat. 176, 377–386 (2019).
Giordano, S. H. Breast cancer in men. N. Engl. J. Med. 379, 1385–1386 (2018).
Wang, F. et al. Sex disparity observed for oncotype DX breast recurrence score in predicting mortality among patients with early stage ER-positive breast cancer. Clin. Cancer Res. 26, 101–109 (2020).
Massarweh, S. A. et al. Molecular characterization and mortality from breast cancer in men. J. Clin. Oncol. 36, 1396–1404 (2018).
Sanchez-Munoz, A. et al. Male breast cancer: correlation between immunohistochemical subtyping and PAM50 intrinsic subtypes, and the subsequent clinical outcomes. Mod. Pathol. 31, 299–306 (2018).
Bayani, J. et al. Molecular stratification of early breast cancer identifies drug targets to drive stratified medicine. npj Breast Cancer 3, 3 (2017).
Bartlett, J. M. S. et al. Computational approaches to support comparative analysis of multiparametric tests: modelling versus training. PLoS ONE 15, e0238593–e0238593 (2020).
Cardoso, F. et al. Characterization of male breast cancer: results of the EORTC 10085/TBCRC/BIG/NABCG International Male Breast Cancer Program. Ann. Oncol. 29, 405–417 (2018).
Bartlett, J. M. et al. Comparing breast cancer multiparameter tests in the OPTIMA prelim trial: no test is more equal than the others. J. Natl. Cancer Inst. 108, djw050 (2016).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Eng. J. Med. 351, 2817–2826 (2004).
Paik, S. et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. Journal of Clinical Oncology 24, 3726–3734 (2006).
Chia, S. K. et al. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin. Cancer Res. 18, 4465–4472 (2012).
Nielsen, T. O. et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin. Cancer Res. 16, 5222–5232 (2010).
Cardoso, F. et al. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. New Eng. J. Med. 375, 717–729 (2016).
van’t Veer, L. J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002).
van de Vijver, M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl. J. Med. 347, 1999–2009 (2002).
Ma, X. J. et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clinical Cancer Res. 14, 2601–2608 (2008).
Dowsett, M. et al. Predictive algorithms for adjuvant therapy: TransATAC. Steroids 76, 777–780 (2011).
Sestak, I. et al. Comparison of the performance of 6 Prognostic signatures for estrogen receptor–positive breast cancer: a secondary analysis of a randomized clinical trialprognostic signatures for estrogen receptor–positive breast cancerprognostic signatures for estrogen receptor–positive breast cancer. JAMA Oncol. 4, 545–553 (2018).
Leyland-Jones, B. R. et al. Recommendations for collection and handling of specimens from group breast cancer clinical trials. J. Clin. Oncol. 26, 5638–5644 (2008).
Sparano, J. A. et al. Clinical and genomic risk to guide the use of adjuvant therapy for breast cancer. N. Eng. J. Med. 380, 2395–2405 (2019).
Cuzick, J. et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the genomic health recurrence score in early breast cancer. J. Clin. Oncol. 29, 4273–4278 (2011).
Blanche, P., Dartigues, J. F. & Jacqmin-Gadda, H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 32, 5381–5397 (2013).
Bayani, J. et al. Metadata record for the article: Evaluation of multiple transcriptomic gene risk signatures in male breast cancer. https://doi.org/10.6084/m9.figshare.14616831(2021).
Acknowledgements
The funding source had no role in study design or conduct, data collection, data management, data analysis, data interpretation, or writing of the report. This work has been funded by the Breast Cancer Research Foundation (BCRF) with additional funding provided by the Government of Ontario to the Ontario Institute of Cancer Research (OICR). This work was funded by the Breast Cancer Research Foundation (BCRF), the Susan G. Komen for the Cure Foundation and the Ontario Institute of Cancer Research (OICR). Funding for OICR is provided by the Government of Ontario.
Author information
Authors and Affiliations
Contributions
All authors approved the submitted version and are personally accountable for their own contributions. Jane Bayani, Coralie Poncet, Cheryl Crozier, Anouk Neven, Fatima Cardoso, and John Bartlett were responsible for the conception and design of the study, the acquisition, analysis, and interpretation of data and have collectively drafted and revised the study for publication. Tammy Piper, Carrie Cunningham, Monika Sobol were responsible for the acquisition and analysis of data and provided revisions of the study for publication. Stefan Aebi, Kim Benstead, Oliver Bogler, Lissandra Dal Lago, Judith Fraser, Florentine Hilbers, Ingrid Hedenfalk, Larissa Korde, Barbro Linderholm, John Martens, Lavinia Middleton, Melissa Murray, Catherine Kelly, Cecilia Nilsson, Monika Nowaczyk, Stephanie Peeters, Aleksandra Peric, Peggy Porter, Carolien Schröder, Isabel T. Rubio, Kathryn J. Ruddy, Christi van Asperen, Danielle Van Den Weyngaert, Carolien van Deurzen, Elise van Leeuwen-Stok, Joanna Vermeij, Eric Winer, and Sharon H. Giordano were responsible for the acquisition, analysis and interpretation of data and provided revisions of the study for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests, no other competing interests have been declared: Dr. John Bartlett has received consultancies/honoraria from Insight Genetics, Inc., BioNTech AG, Biotheranostics, Inc., Pfizer, Rna Diagnostics Inc., oncoXchange/MedcomXchange Communications Inc, OncoCyte Corporation, NanoString Technologies, Inc., Oncology Education. Research funding from ThermoFisher Scientific, Genoptix, Agendia, NanoString Technologies, Inc., Stratifyer GmbH and Biotheranostics, Inc. Dr. Bartlett has received travel support from Biotheranostics, Inc., NanoString Technologies, Inc. and the Breast Cancer Society of Canada. Dr. Bartlett and Dr. Barbro Linderholm are members of advisory boards for Astra Zeneca, Pfizer, Eli Lilly, Novartis, Merck, and Daiichi Sankyo. Dr. Jane Bayani is a co-applicant on multiple patents including those for breast cancer prognostic signatures.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bayani, J., Poncet, C., Crozier, C. et al. Evaluation of multiple transcriptomic gene risk signatures in male breast cancer. npj Breast Cancer 7, 98 (2021). https://doi.org/10.1038/s41523-021-00301-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41523-021-00301-0