Abstract
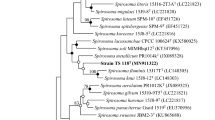
An aerobic, Gram-stain-negative, non-motile, non-spore-forming, rod-shaped, and light pink-colored bacterial strain, designated TS19T, was isolated from a sand sample obtained from a coastal sand dune after exposure to 3 kGy of gamma radiation. Phylogenetic analysis based on the 16S rRNA gene sequences revealed that the isolate was a member of the genus Hymenobacter and was most closely related to H. wooponensis WM78T (98.3% similarity). Strain TS19T and H. wooponensis showed resistance to gamma radiation with D10 values (i.e., the dose required to reduce the bacterial population by tenfold) of 7.3 kGy and 3.5 kGy, respectively. The genome of strain TS19T consists of one contig with 4,879,662 bp and has a G + C content of 56.2%. The genome contains 3,955 protein coding sequences, 44 tRNAs, and 12 rRNAs. The predominant fatty acids of strain TS19T were iso-C15:0, summed feature 4 (iso-C17:1 I and/or anteiso-C17:1 B), summed feature 3 (C16:1 ω6c and/or C16:1 ω7c), and C16:1 ω5c. The major polar lipids were phosphatidylethanolamine, and one unidentified aminophospholipid. The main respiratory quinone was menaquinone-7. Based on the phylogenetic, physiological, and chemotaxonomic characteristics, strain TS19T represents a novel species, for which the name Hymenobacter taeanensis sp. nov. is proposed. The type strain is TS19T (= KCTC 72897T = JCM 34023T).


Similar content being viewed by others
Availability of data and materials
The complete genome sequence of strain TS19T generated during the current study has been deposited in GenBank with the accession codes, https://ncbi.nlm.nih.gov/nuccore/ NZ_CP053538. The 16S rRNA gene sequence also is available in the GenBank repository, https://ncbi.nlm.nih.gov/nuccore/MN911321.
References
Bernardet JF, Nakagawa Y, Holmes B, Bernardet JF, Nakagawa Y, Holmes B, Subcommittee On The Taxonomy Of F, Cytophaga Like Bacteria Of The International Committee On Systematics Of P (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Buczolits S, Denner EBM, Kampfer P, Busse HJ (2006) Proposal of Hymenobacter norwichensis sp. nov., classification of 'Taxeobacter ocellatus’, 'Taxeobacter gelupurpurascens’ and 'Taxeobacter chitinovorans’ as Hymenobacter ocellatus sp. nov., Hymenobacter gelipurpurascens sp. nov. and Hymenobacter chitinivorans sp. nov., respectively, and emended description of the genus Hymenobacter Hirsch et al. 1999. Int J Syst Evol Microbiol 56:2071–2078
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569
Chun J, Goodfellow M (1995) A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol 45:240–245
Collins MD (1985) Analysis of isoprenoid quinones. Methods Microbiol 18:329–366
Collins MD, Hutson RA, Grant IR, Patterson MF (2000) Phylogenetic characterization of a novel radiation-resistant bacterium from irradiated pork: description of Hymenobacter actinosclerus sp. nov. Int J Syst Evol Microbiol 50(Pt 2):731–734
Da Costa MS, Albuquerque L, Nobre MF, Wait R (2011) The identification of polar lipids in prokaryotes. Methods Microbiol 38:165–181
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Felsenstein J (1993) PHYLIP (phylogeny inference package), version 3.5 c. Joseph Felsenstein
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416
Gabani P, Singh OV (2013) Radiation-resistant extremophiles and their potential in biotechnology and therapeutics. Appl Microbiol Biotechnol 97:993–1004
Gerber E, Bernard R, Castang S, Chabot N, Coze F, Dreux-Zigha A, Hauser E, Hivin P, Joseph P, Lazarelli C (2015) Deinococcus as new chassis for industrial biotechnology: biology, physiology and tools. J Appl Microbiol 119:1–10
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Han L, Wu SJ, Qin CY, Zhu YH, Lu ZQ, Xie B, Lv J (2014) Hymenobacter qilianensis sp. nov., isolated from a subsurface sandstone sediment in the permafrost region of Qilian Mountains, China and emended description of the genus Hymenobacter. Antonie Van Leeuwenhoek 105:971–978
Hirsch P, Ludwig W, Hethke C, Sittig M, Hoffmann B, Gallikowski CA (1998) Hymenobacter roseosalivarius gen. nov., sp. nov. from continental Antartica soils and sandstone: bacteria of the Cytophaga/Flavobacterium/Bacteroides line of phylogenetic descent. Syst Appl Microbiol 21:374–383
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286-293
Jin M, Xiao A, Zhu L, Zhang Z, Huang H, Jiang L (2019) The diversity and commonalities of the radiation-resistance mechanisms of Deinococcus and its up-to-date applications. AMB Express 9:1–12
Kang JW, Lee JH, Baik KS, Lee SS, Seong CN (2015) Hymenobacter wooponensis sp. nov., isolated from wetland freshwater. Int J Syst Evol Microbiol 65:1871–1876
Kim KH, Im WT, Lee ST (2008) Hymenobacter soli sp. nov., isolated from grass soil. Int J Syst Evol Microbiol 58:941–945
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Klassen JL, Foght JM (2008) Differences in carotenoid composition among hymenobacter and related strains support a tree-like model of carotenoid evolution. Appl Environ Microbiol 74:2016–2022
Klassen JL, Foght JM (2011) Characterization of Hymenobacter isolates from Victoria Upper Glacier, Antarctica reveals five new species and substantial non-vertical evolution within this genus. Extremophiles 15:45–57
Kovacs N (1956) Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178:703
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee JJ, Park SJ, Lee YH, Lee SY, Ten LN, Jung HY (2017) Hymenobacter aquaticus sp. nov., a radiation-resistant bacterium isolated from a river. Int J Syst Evol Microbiol 67:1206–1211
Lee SE, Ten LN, Park Y, Maeng S, Zhang J, Kim M-K, Cha I-T, Lee K-E, Lee B-H, Jung H-Y, Kim MK (2021) Hymenobacter busanensis sp. nov., radiation-resistant species isolated from soil in South Korea. Arch Microbiol 203:755–762
Lim S, Jung JH, Blanchard L, de Groot A (2019) Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol Rev 43:19–52
Maeng S, Kim MK, Subramani G (2020) Hymenobacter jejuensis sp. nov., a UV radiation-tolerant bacterium isolated from Jeju Island. Antonie Van Leeuwenhoek 113:553–561
Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Munoz R, Rossello-Mora R, Amann R (2016) Revised phylogeny of Bacteroidetes and proposal of sixteen new taxa and two new combinations including Rhodothermaeota phyl. nov. Syst Appl Microbiol 39:281–296
Orellana R, Macaya C, Bravo G, Dorochesi F, Cumsille A, Valencia R, Rojas C, Seeger M (2018) Living at the frontiers of life: extremophiles in Chile and their potential for bioremediation. Front Microbiol 9:2309
Powers EM (1995) Efficacy of the Ryu nonstaining KOH technique for rapidly determining gram reactions of food-borne and waterborne bacteria and yeasts. Appl Environ Microbiol 61:3756–3758
Reddy G (2013) Phylogenetic analyses of the genus Hymenobacter and description of Siccationidurans gen. nov., and Parahymenobacter gen. nov. J Phylogen Evolution Biol 1:2
Reddy G, Garcia-Pichel F (2013) Description of Hymenobacter arizonensis sp. nov. from the southwestern arid lands of the United States of America. Antonie Van Leeuwenhoek 103:321–330
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. MIDI inc, Newark
Sedláček I, Pantůček R, Holochová P, Králová S, Staňková E, Vrbovská V, Šedo O, Švec P, Busse HJ (2019a) Hymenobacter humicola sp. nov., isolated from soils in Antarctica. Int J Syst Evol Microbiol 69:2755–2761
Sedláček I, Pantůček R, Králová S, Mašlaňová I, Holochová P, Staňková E, Vrbovská V, Švec P, Hans-Jürgen Busse H-J (2019b) Hymenobacter amundsenii sp. nov. resistant to ultraviolet radiation, isolated from regoliths in Antarctica. Syst Appl Microbiol 42:284–290
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Smibert R, Krieg N, Gerhardt P, Murray R, Wood W (1994) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–654
Su S, Chen M, Teng C, Jiang S, Zhang C, Lin M, Zhang W (2014) Hymenobacter kanuolensis sp. nov., a novel radiation-resistant bacterium. Int J Syst Evol Microbiol 64:2108–2112
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Timmins J, Moe E (2016) A decade of biochemical and structural studies of the DNA repair machinery of Deinococcus radiodurans: major findings, functional and mechanistic insight and challenges. Comput Struct Biotechnol J 14:168–176
Tittsler RP, Sandholzer LA (1936) The use of semi-solid agar for the detection of bacterial motility. J Bacteriol 31:575–580
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Funding
This research was supported by the Nuclear R&D program of the Ministry of Science and ICT (MSIT), Republic of Korea.
Author information
Authors and Affiliations
Contributions
Conceptualization, funding acquisition, and supervision: SL; Resources: M-KK; Investigation validation, and visualization: JHL, J-HJ, HNC; Data curation and formal analysis: JHL, J-HJ; Writing-original draft: JHL; Writing-review and editing: SL.
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflicts of interest declare that are relevant to the content of this article.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain TS19T is MN911321.
The GenBank/EMBL/DDBJ accession number for the whole genome sequence of strain TS19T is NZ_CP053538.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, J.H., Jung, JH., Kim, MK. et al. Hymenobacter taeanensis sp. nov., radiation resistant bacterium isolated from coastal sand dune. Antonie van Leeuwenhoek 114, 1585–1593 (2021). https://doi.org/10.1007/s10482-021-01624-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01624-5