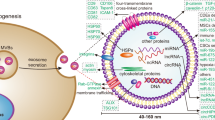
Abstract
Exosomal microRNAs (miRNAs) have been implicated in the development and progression of a variety of tumors; however, whether they contribute to medulloblastoma (MB) tumorigenesis remains to be elucidated. To address this, we first characterized the miRNA profiles of circulating exosomes by miRNA sequencing to identify miRNAs differentially expressed between children with MB and healthy controls. Then, we conducted in vitro and in vivo functional assays with the identified miRNAs and their predicted targets. We found that, compared with healthy controls, 35 miRNAs were upregulated and 5 downregulated in exosomes isolated from the plasma of MB patients. We further found that the expression of miR-101-3p and miR-423-5p was significantly higher in plasma exosomes from MB patients than in healthy controls in an expanded cohort and these exosomal miRNAs could be delivered to tumor cells via exosomes. An in vitro functional analysis of miR-101-3p and miR-423-5p showed that treating MB cells with the corresponding mimics significantly inhibited the proliferation, colony-forming ability, migratory ability, and invasive capacity of tumor cells, and promoted cell apoptosis. Additionally, miR-101-3p and miR-423-5p were found to act as tumor suppressors by directly targeting a common gene, FOXP4, which encodes a transcription factor with a vital role in embryonic development and tumorigenesis. Moreover, miR-101-3p also targeted EZH2, a histone methyltransferase, to reinforce its tumor inhibitory effects. Using a xenograft nude mouse model of MB, we further identified that the overexpression of miR-101-3p and miR-423-5p inhibited tumorigenesis in vivo. Our findings provide novel insights into the functions of exosomal miRNAs in mediating MB progression and suggest a potential therapeutic approach for the treatment of children with MB.
Similar content being viewed by others
Introduction
Medulloblastoma (MB) is the most commonly diagnosed malignant brain tumor in children. The 5-year overall survival rate for patients with MB is approximately 60–70% under the current standard multimodal treatment, which consists of maximal safe tumor resection, chemotherapy, and radiotherapy. Survivors often experience serious long-term sequelae resulting from these aggressive therapies, including neurocognitive impairment, endocrine deficiencies, and even secondary tumors [1, 2]. Therefore, more effective, less toxic therapies are urgently needed to improve the outcomes and quality of life for MB patients.
Exosomes are lipid bilayer-enclosed vesicles, 30–150 nm in diameter, that are actively secreted by many cell types. Exosomes are important for cell communication as carriers of a variety of biologically active molecules, including proteins, lipids, mRNAs, and microRNAs (miRNAs) [3, 4]. They are involved in normal physiological processes, but also have important pathophysiological roles, including in cardiovascular diseases, inflammation, neurodegenerative diseases, and tumor development [5, 6]. Several reliable methods currently exist for the isolation of exosomes from biofluids, including blood, urine, milk, and cell culture medium. Although differential ultracentrifugation is the most widely used of these methods, other techniques represent credible alternatives, including ultrafiltration, the use of precipitating agents such as polyethylene glycol, immunoaffinity capture, microfluidics, and size-exclusion chromatography [7]. MiRNAs are small, non-coding RNAs approximately 19–22 nucleotides long that regulate gene expression either through translational repression or by directly targeting mRNAs for degradation, and play vital roles in nearly all biological processes [8]. Exosomal miRNAs can serve as important intercellular communication tools and exosome-mediated transfer of oncogenic or tumor-suppressor miRNAs can influence tumorigenesis. For instance, in metastatic lung cancer cell lines, the exosomal miRNA, miR-21, induces VEGF production via STAT3, thereby promoting tumor angiogenesis and metastasis [9]. Tumor-specific, exosome-derived miRNAs have been proposed as potential markers for lung, nasopharyngeal, ovarian, and prostate cancers, as well as other malignant tumors that affect adults [10,11,12,13,14]. Munoz et al. [15] found that mesenchymal stem cell-derived exosomes could deliver miR-9 antagonists to glioblastoma cells, thereby blocking the expression of miR-9 and increasing glioblastoma sensitivity to various treatments.
Multiple tumor-resident miRNAs have been reported to be involved in the progression and development of brain tumors in children, including central nervous system primitive neuroectodermal brain tumors, pediatric pilocytic astrocytoma, ependymal tumors, and MBs [5]. MiR-218, miR-19a, miR-20, and miR-92 were found to be aberrantly expressed in human MBs, affecting multiple tumorigenic processes, including cell-cycle progression, differentiation, and apoptosis, through the regulation of critical signaling pathways such as the Wnt/β-catenin and sonic hedgehog pathways [16, 17]. However, whether exosomal-derived miRNAs contribute to MB tumorigenesis remains to be elucidated.
We have previously reported that exosomal miR-130b-3p functions as a tumor suppressor in MB both in vitro and in vivo by targeting serine/threonine-protein kinase 1 (SIK1) through p53-related pathways [18]. Based on different selection criteria and larger screening samples, we report here that the plasma levels of exosomal miR-101-3p and miR-423-5p are also upregulated in MB patients when compared with those of healthy controls. MiR-101-3p and miR-423-5p were found to exert antitumorigenic effects both in vitro and in vivo by targeting FOXP4 and EZH2, both of which were upregulated in MB tissues. Together, our findings demonstrated that exosomal miR-101-3p and miR-423-5p have previously unrecognized roles in the pathogenesis of childhood MB and are promising therapeutic targets for the treatment of this disease.
Results
MiR-101-3p and miR-423-5p levels are upregulated in exosomes derived from the plasma of MB patients, and these miRNAs can be transferred to tumor cells via exosomes
In preliminary screening, exosomes were isolated from the plasma of MB patients (n = 4) and age-matched healthy controls (n = 4) using ultracentrifugation. Isolated exosomes were analyzed by transmission electron microscopy to determine the average exosome size and morphology, revealing typical double-layered spherical structures with diameters <150 nm (Fig. 1A). To further validate our exosomal preparations, we detected the expression of the exosomal markers CD9, CD63, and GM130 by western blot. The isolated particles were positive for the exosomal core protein markers, CD9 and CD63, and negative for the cis-Golgi marker, GM130 (Fig. 1B). The exosomes were further analyzed by nanoparticle tracking analysis (NTA) and found to be predominantly in the 50–150-nm size range in both MB patients and normal controls (Fig. S1A, B). No significant differences in exosome concentrations were found between the two groups (Fig. S1C).
A Transmission electron microscopy analysis. Scale bar, 100 nm. B Western blotting analysis of the exosomal marker proteins CD9, CD63, and GM130. C Heatmap showing exosomal miRNAs differentially expressed between medulloblastoma (MB) patients and healthy controls (fold change >1.5). D Heatmap showing exosomal miRNAs differentially expressed at fold change >1.5 and p < 0.05. E, F RT-qPCR-based verification of miR-101-3p and miR-423-5p expression levels in plasma-derived exosomes (E) or peripheral blood mononuclear cells (PBMCs) (F) in an expanded cohort (MB = 35, control = 17). G Internalization of plasma-derived exosomes into Daoy cells. Scale bar indicates 50 µm. H The expression levels of miR-101-3p and miR-423-5p in Daoy cells after culture with plasma-derived exosomes isolated from MB patients and normal controls. Data are presented as means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we investigated the miRNA expression profiles of plasma-derived exosomes by miRNA-seq. Using a 1.5-fold change as the threshold to define up- or downregulated miRNAs, we found 35 miRNAs that were upregulated and 5 that were downregulated in MB patients when compared with healthy controls (Fig. 1C). We then selected a p value < 0.05 to further narrow the range of differentially expressed miRNAs and found that the expression of miR-101-3p, miR-320b-3p, miR-20a-5p, and miR-423-5p was higher in exosomes derived from the plasma of MB patients than in those derived from the plasma of healthy controls (Fig. 1D). We then examined the expression levels of the differentially expressed miRNAs in an expanded cohort by quantitative real-time PCR (qPCR). The expression of miR-101-3p and miR-423-5p was upregulated in exosomes derived from the plasma of MB patients relative to that of healthy controls (Fig. 1E). We further investigated the expression of these two miRNAs according to four known molecular MB subgroups (WNT, SHH, Group 3, and Group 4) and found no significant differences in miR-101-3p and miR-423-5p expression levels among these subgroups (Fig. S2A, B). We also compared the expression of miR-101-3p and miR-423-5p in peripheral blood mononuclear cells (PBMCs) from MB patients and healthy controls and found that the expression of both miRNAs was higher in the PBMCs of MB patients (Fig. 1F).
To determine whether plasma-derived exosomes could be transferred to recipient tumor cells, we cultured Daoy or D283 Med (MB cell lines) cells with pooled PKH67-labeled exosomes isolated from MB patients for 24 h. Subsequent confocal microscopy analysis confirmed the presence of PKH67-labeled exosomes in the cytoplasm of the cancer cells, implying that peripheral blood-derived exosomes can be internalized by tumor cells (Fig. 1G and Fig. S3A) . Consistent with this observation, we found that the expression levels of miR-101-3p and miR-423-5p were upregulated in both Daoy and D283 Med cells after culture with pooled exosomes derived from the plasma of MB patients, whereas no significant change was observed following culture with exosomes derived from the plasma of healthy controls (Fig. 1H and Fig. S3B). Together, these results showed that miR-101-3p and miR-423-5p were enriched in exosomes derived from the plasma of MB patients and could be delivered to tumor cells via these exosomes.
MiR-101-3p and miR-423-5p function as tumor suppressors in MB cells in vitro
We next analyzed the effects of miR-101-3p and miR-423-5p on the proliferative capacity of MB cells using a Cell Counting Kit-8 (CCK-8) assay. The overexpression of miR-101-3p or miR-423-5p in Daoy and D283 Med cells resulted in decreased tumor cell proliferation rates compared with those of the controls (Fig. 2A, B). We further assessed the effects of adding exosomes (50 μg/ml) derived from the plasma of MB patients on the proliferation of Daoy and D283 Med cells by CCK-8 assay, with the results showing that the proliferative ability of tumor cells from both cell lines was significantly inhibited (Fig. 2C). Next, we investigated the effect of miR-101-3p and miR-423-5p on cell apoptosis via FITC–Annexin V/propidium iodide double staining, followed by flow cytometric analysis. The overexpression of either miRNA led to a significant increase in the rate of apoptosis of Daoy and D283 Med cells compared with that in the negative controls (Fig. 2D, E).
A, B Cell Counting Kit-8 (CCK-8) assays showing that Daoy and D283 Med cells transfected with miR-101-3p/miR-423-5p mimics or lentivirus exhibit decreased cell proliferation compared with cells transfected with miR-NC (negative control). C A CCK-8 cell proliferation assay was performed on Daoy and D283 Med cells cultured with pooled exosomes (50 µg/ml) derived from the plasma of medulloblastoma patients. D, E Flow cytometric analysis of cell apoptosis in Daoy and D283 Med cells transfected with miR-101-3p or miR-423-5p mimics or lentiviral vectors. F MiR-101-3p and miR-423-5p mimics inhibited Daoy cell migration into wound areas. Scale bar indicates 100 µm. G, H Transwell assays showing that the migratory and invasive abilities of Daoy cells transfected with miR-101-3p or miR-423-5p mimics are inhibited compared with those of cells transfected with miR-NC. Scale bar indicates 100 µm. Data are presented as means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Migration and Matrigel invasion assays were also performed to assess the effects of miR-101-3p and miR-423-5p on cell motility. The overexpression of either miR-101-3p or miR-423-5p inhibited the invasive and migratory abilities of Daoy cells (Fig. 2F–H). Taken together, these data confirmed that both miR-101-3p and miR-423-5p exert antitumorigenic effects in MB cells, and that the upregulation of either miRNA can inhibit MB cell proliferation, migration, and invasion, while also promoting cell apoptosis.
Monocyte/macrophage-derived exosomal miR-101-3p and miR-423-5p can be transferred into MB cells
We found that miR-101-3p and miR-423-5p were both upregulated in PBMCs and exosomes isolated from the plasma of MB patients (Fig. 1E, F), whereas the expression of both miRNAs was lower in tumor tissues than in adjacent normal brain tissues (Fig. 3A). This suggested that the exosomes containing these two miRNAs were derived from immune cells rather than tumor cells as a result of the immune response against the tumors. Macrophage-derived exosomes account for a large proportion of circulating microvesicles in blood [19]. To determine the origin of the exosomes containing miR-101-3p and miR-423-5p, we isolated exosomes from THP-1 monocyte culture supernatants and detected the expression of miR-101-3p and miR-423-5p. We found that the expression of miR-101-3p and miR-423-5p was higher in the isolated exosomes than the THP-1 cells (Fig. 3B). Then, we cultured Daoy cells with the exosomes derived from the THP-1 cells transfected with miR-101-3p/miR-423-5p mimics or miR-NC, and observed that these exosomes could also be transferred into Daoy cells and that the expression levels of miR-101-3p and miR-423-5p were higher after culture (Fig. 3C, D). The proliferative ability of Daoy cells was significantly inhibited by the exosomes derived from the supernatant of THP-1 cells transfected with miR-101/miR-423 mimics compared with controls (Fig. 3E).
A The expression levels of miR-101-3p and miR-423-5p in tumor tissues from medulloblastoma (MB) patients relative to those in adjacent normal brain tissues (MB = 25, control = 15). B The expression of miR-101-3p and miR-423-5p in THP-1 cells and exosomes derived from THP-1 cell culture supernatants. C Exosomes from THP-1 cell culture supernatants cultured with Daoy cells. Scale bar indicates 50 µm. D The expression of miR-101-3p and miR-423-5p in Daoy cells cultured with THP-1-derived exosomes. E The proliferative ability of Daoy cells cultured with exosomes (50 µg/ml) derived from supernatant of THP-1 cells transfected with miR-101-3p/miR-423-5p or miR-NC (negative control). F The proliferative ability of Daoy cells cultured with supernatant of THP-1 cells transfected with miR-101-3p/miR-423-5p mimics or miR-NC (negative control) and treated or not with GW4869 (40 µM). G Immunohistochemical analysis of CD68 expression in MB tissue. Scale bar indicates 100 µm. H Compared with that in the controls, the expression of exosomal miR-101-3p and miR-423-5p was higher in MB patient-derived CD14+ monocyte culture supernatant. Data are presented as means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
We further cultured Daoy cells with culture supernatants of THP-1 monocytes transfected with miR-101-3p/miR-423-5p mimics or miR-NC (negative control) and found that the proliferative ability of Daoy cells was significantly inhibited, whereas no significant change was observed following culture with medium containing GW4869 (40 μM; a neutral sphingomyelinase inhibitor that inhibits exosome secretion) (Fig. 3F). These results indicated that the effects on cell proliferation were at least partly due to the presence of exosomes, and that these effects could be partially reversed by GW4869 treatment. Similar to THP-1 cells, we found that HMO6 cells (a type of glial cell equivalent to a macrophage in the brain) could also secrete exosomes containing miR-101-3p and miR-423-5p (Fig. S4A–D) and CD68+ cells exist in MB tissues (Fig. 3G). Finally, we isolated CD14+ monocytes from the peripheral blood of MB patients and healthy controls and cultured them for 72 h. Compared with that of controls, the expression of exosomal miR-101-3p and miR-423-5p was higher in culture supernatants of MB patient-derived CD14+ monocytes (Fig. 3H). Together, these results suggested that monocyte-macrophages in MB patients secreted exosomes containing miR-101-3p and miR-423-5p, which could be transferred to MB cells.
FOXP4 is a target for both miR-101-3p and miR-423-5p
To further understand how miR-101-3p and miR-423-5p influence tumor progression, we identified their potential targets using TargetScan and RNA22 prediction tools. Six genes predicted to be regulated by both miRNAs in the two databases were chosen for evaluation as potential targets, namely FOXP4, PLCB1, ANKRD52, ANGPTL2, DLGAP3, and DCUN1D3 (Fig. 4A). Because FOXP4 (forkhead box P4) is known to have roles in embryonic development and tumorigenesis [20], this gene was selected for further study. Daoy cells transfected with either miR-101-3p or miR-423-5p displayed reduced expression of endogenous FOXP4 protein as determined by western blotting (Fig. 4B, C). Furthermore, luciferase reporter assays in HEK293T cells showed that the overexpression of either miRNA markedly suppressed FOXP4-3′UTR-driven luciferase activity; however, no significant change in luciferase activity was observed following the co-transfection of miR-101-3p or miR-423-5p with a mutated FOXP4 3′UTR (Fig. 4D). This indicated that FOXP4 may be a direct and functional target of both miR-101-3p and miR-423-5p in MB.
A A Venn diagram showing target genes shared between miR-101-3p and miR-423-5p, as predicted from two databases (TargetScan and RNA22). B Western blot analysis of FOXP4 protein expression in Daoy cells transfected with the miR-101-3p mimic or miR-NC (negative control). C Western blot analysis of FOXP4 protein expression in Daoy cells transfected with the miR-423-5p mimic or miR-NC. D The predicted binding sites of miR-101-3p and miR-423-5p in the FOXP4- wild or mutated 3′UTR (left). Dual-luciferase assay in HEK293T cells co-transfected with the miR-101-3p or miR-423-5p mimic and a construct containing the wild-type or mutated FOXP4-3′UTR (right). Data are presented as means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
FOXP4 functions as an oncogene in MB cells
We first evaluated the expression levels of FOXP4 in MB tumor tissues by real-time qPCR. The expression levels of FOXP4 were higher in MB tumor tissues than in adjacent brain tissue samples (Fig. 5A). To further clarify whether targeting FOXP4 could mediate MB tumor progression, we performed a series of functional assays in Daoy cells. Inhibiting FOXP4 expression using siRNA (Fig. 5B) resulted in reduced cell proliferation, migration, and invasion, whereas the rates of cell apoptosis were increased (Fig. 5C–H). These results strongly indicated that silencing FOXP4 phenocopied the inhibitory effects of miR-101-3p and miR-423-5p overexpression on tumor progression. Combined, these results revealed that FOXP4 may function as an oncogene in MB tumorigenesis and may be a target for both miR-101-3p and miR-423-5p.
A Comparison of the expression levels of FOXP4 in tumor samples from medulloblastoma (MB) patients and those in adjacent normal tissues as determined by qPCR (MB = 25, control = 15). B The knockdown of FOXP4 in Daoy cells using siRNAs; siFOXP4-1001 was selected for use in subsequent experiments. C, D The knockdown of FOXP4 inhibited cell proliferation and clonogenicity in Daoy cells. E Wound-healing assay showing that knockdown of FOXP4 using siFOXP4-1001 inhibited cell migration into wound areas. Scale bar indicates 100 µm. F, G Transwell assays showing that the knockdown of FOXP4 decreased cell migration and invasion Scale bar indicates 100 µm. H The knockdown of FOXP4 promoted cell apoptosis, as determined by flow cytometry. Data are presented as means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
EZH2 is upregulated in MB tumor tissues, is a functional target of miR-101-3p, and acts as an oncogene in MB tumor progression
We observed that, in vitro, the inhibitory effects of miR-101-3p on Daoy cells appeared to be stronger than those of miR-423-5p (Fig. 2A). This suggested that miR-101-3p may also target genes other than FOXP4. Through bioinformatics analysis, we identified EZH2 (enhancer of zeste homolog 2) as another putative target of miR-101-3p. The EZH2 protein, a histone methyltransferase, is highly expressed in several malignancies and is responsible for the H3K27me3 modification in the promoter regions of target genes. The latter is pivotal for various biological processes, including differentiation, the maintenance of cell identity, and proliferation [21, 22]. EZH2 expression is increased in MB and the genetic or pharmacological targeting of EZH2 impairs MB cell growth [23].
As shown in Fig. 6A, the relative expression of EZH2 mRNA was significantly higher in tumor tissue than in adjacent normal tissue. We also investigated whether there were seed-matching sites for miR-101-3p in the EZH2 3′UTR using the TargetScan database, and employed a dual-luciferase reporter assay to confirm the correlation between miR-101-3p and EZH2 expression levels. As shown in Fig. 6B, the transfection of the miR-101-3p mimic decreased the luciferase activity driven by the wild-type EZH2 3′UTR, whereas no significant change was seen following transfection with a mutated form, suggesting that miR-101-3p can inhibit the expression of EZH2 by directly binding to sites in its 3′UTR. Transient transfection of miR-101-3p also reduced EZH2 mRNA and protein levels in Daoy cells (Fig. 6C, D).
A Relative EZH2 mRNA expression in tumor samples (n = 25) compared with that in normal tissues (n = 15). B Predicted binding site of miR-101-3p in the EZH2- wild or mutated 3′UTR (left). Luciferase reporter assay in HEK293T cells co-transfected with the miR-101-3p mimic and a construct containing the wild-type or mutated EZH2 3′UTR (right). C EZH2 mRNA expression in Daoy cells transfected with the miR-101-3p mimic. D EZH2 protein level in Daoy cells transfected with the miR-101-3p mimic. E Knockdown of EZH2 by siRNAs; siEZH2-614 was selected for use in subsequent experiments. F–G Cell proliferation and clonogenicity were significantly inhibited in Daoy cells transfected with siEZH2. H The knockdown of EZH2 significantly inhibited cell migration into wound areas in Daoy cells. Scale bar indicates 100 µm. I, J Transwell assays showing that migratory and invasive abilities are inhibited in Daoy cells transfected with siEZH2. Scale bar indicates 100 µm. K Flow cytometry results showing that cell apoptosis rates were increased in Daoy cells transfected with siEZH2. Data are presented as means ± SEM from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Next, to investigate the function of EZH2 in MB, we knocked down its expression in Daoy cells using siRNA (Fig. 6E). Functional assays showed that the downregulation of EZH2 inhibited MB cell viability, migration, and invasion, while promoting MB cell apoptosis (Fig. 6F–K). Taken together, these results demonstrated that miR-101-3p can inhibit MB by directly targeting EZH2, as well as FOXP4.
The overexpression of miR-101-3p or miR-423-5p inhibits tumorigenesis in vivo
To evaluate the antitumorigenic effects of miR-101-3p and miR-423-5p in vivo, we subcutaneously injected Daoy cells (5 × 106) transfected with LV16-miR-101-3p, LV16-miR-423-5p, or LV16-NC (negative control) into male Balb/c nude mice and measured the volumes of the resulting tumors weekly. The relative expression levels of miR-101-3p and miR-423-5p in Daoy cells transfected with lentivirus were measured by RT-qPCR (Fig. 7A). Tumors became palpable 2 weeks postinjection and the mice were sacrificed at 7 weeks post-implantation (Fig. 7B, C). Tumor growth was significantly suppressed in the LV16-miR-101-3p- and LV16-miR-423-5p-treated groups relative to the control group (Fig. 7C, D). As shown in Fig. 7E, there was also a marked reduction in tumor weight following treatment with either miR-101-3p or miR-423-5p. We also evaluated the expression of EZH2, FOXP4, and Ki-67 in xenograft tumor tissues by immunohistochemistry. The levels of FOXP4 and Ki-67 were both downregulated in the LV16-miR-101-3p- and LV16-miR-423-5p-expressing tumor tissues compared with those in control tissues (Fig. 7F, G), while EZH2 was also downregulated in the LV16-miR-101-3p group. Together, these data suggested that miR-101-3p and miR-423-5p inhibit tumor growth in vivo by inhibiting the expression of FOXP4 and EZH2.
A Relative expression levels of miR-101-3p and miR-423-5p in Daoy cells transfected with LV16-miR-101-3p or LV16-miR-423-5p compared with those in cells transfected with LV16-NC (negative control). B Daoy cells transfected with LV16-miR-101-3p, LV16-miR-423-5p, or LV16-NC were subcutaneously injected into Balb/c nude mice and tumor volumes were measured weekly (n = 6/group/experiment). C Images of tumors (LV16-miR-101-3p/LV16-miR-423-5p vs. LV16-NC). D Growth curves of tumor volumes in the different groups of mice. E Tumor weights of mice at week 7 postinjection. F, G Hematoxylin and eosin staining of tumors and the expression of Ki-67, FOXP4, and EZH2 as determined by immunohistochemistry. Scale bar indicates 100 µm. The animal experiments were performed in duplicate. Data are presented as means ± SEM. *p < 0.05, **p < 0.01.
Discussion
In this study, we demonstrated that the plasma levels of exosomal miR-101-3p and miR-423-5p were significantly higher in MB patients than in healthy controls. Exosomal miR-101-3p and miR-423-5p derived from the plasma of MB patients could be transferred to MB cells and act as tumor suppressors through a common target, FOXP4. These data suggest that exosomal miR-101-3p and miR-423-5p in peripheral blood have potential as therapeutic targets for the treatment of MB patients.
MiR-423-5p is downregulated in malignant colon tissue and colon cancer cell lines [24] and functions as a tumor suppressor in ovarian cancer by inhibiting cell proliferation and invasion [25]; however, miR-423-5p is also highly expressed in glioblastoma tissues and was found to promote cell proliferation, angiogenesis, and invasion through the suppression of its target, ING-4, both in vitro and in vivo [26]. Yang et al. [27] further showed that exosomal miR-423-5p targets SUFU in gastric cancer, thereby promoting cancer growth and metastasis, and can also serve as a marker for this disease. Together, these results indicate that miR-423-5p can function either as a tumor suppressor or as an oncogenic miRNA, depending on its cell-specific targets. In our study, data from both in vitro and in vivo experiments suggested that miR-423-5p functions as a tumor suppressor in MB and has the potential for use as a therapeutic target for the treatment of this disease.
MiR-101 levels are reported to be decreased in human glioblastoma stem cells. The overexpression of miR-101 leads to reduced cell proliferation, migration, and invasion, while also promoting cell apoptosis, by targeting Kruppel-like factor 6 (KLF6), and also inactivates the MEK1/2 and PI3K signaling pathways [28]. Additionally, miR-101 levels are decreased in glioblastoma relative to normal samples, and elevated miR-101 expression is strongly associated with reduced cell growth and motility, both in vivo and in vitro [28]. Here, we found that miR-101-3p functioned as a repressor in MB cells by inhibiting the expression of its newly identified target gene, FOXP4, thereby inhibiting cell proliferation, colony formation, migration, and invasion, while also promoting cell apoptosis.
Mechanistically, our results indicated that FOXP4 is a newly identified common target of miR-101-3p and miR-423-5p in MB, as determined by bioinformatics analysis and luciferase reporter assays. FOXP4, encoding a member of the FOXP subfamily of winged-helix transcription factors, is reported to function as an oncogene in several cancers, including non-small cell lung cancer, hepatocellular carcinoma, and osteosarcoma [29,30,31]. In our study, the expression levels of FOXP4 in tumor tissues were increased when compared with those of normal controls, while the knockdown of FOXP4 impaired tumorigenesis and tumor progression in vitro, suggesting that this protein has an oncogenic role in patients with MB. However, further studies are needed to elucidate the detailed mechanisms underlying the role of FOXP4 in regulating tumorigenesis.
We also found that EZH2 is an unreported target gene of miR-101-3p in MB, in addition to FOXP4. The EZH2 protein is the catalytically active component of polycomb repressor complex 2 (PRC2) and functions as a transcriptional repressor in this context. EZH2 expression is associated with clinical progression, aggressive tumor subgroups, drug resistance, and reduced survival in multiple types of cancers, including cutaneous melanoma, endometrial cancer, prostate cancer, breast cancer, and hepatocellular carcinoma. [32,33,34,35,36] Consistent with these observations, we found that the level of EZH2 expression was higher in MB tumor samples than in normal brain tissues. Hence, our study provides previously unreported insights into the relationship between EZH2 and miR-101-3p in MB, and demonstrates that EZH2 is required for the inhibitory effect of miR-101-3p on tumor progression.
Our results also indicated the potential origin of the exosomes containing miR-101-3p and miR-423-5p. Monocytes can migrate into tissues and differentiate into tissue-resident macrophages, thereby playing an important role in tumor progression and metastasis. Exosome-mediated macrophage reprogramming to the M1 phenotype is reported to have potential as a promising therapy for pancreatic cancer through miR-155 and miR-125b2 transfection using nanoparticle delivery systems [37]. Exosomes from M1-polarized macrophages can enhance the antitumor activity of paclitaxel by activating macrophage-mediated inflammation [38]. Moreover, THP-1- and PBMC-derived macrophages exhibit an M1-like, rather than an M2-like, phenotype when treated with conditioned medium from OSCC cells, but not that from normal epithelium or leukoplakia cells [39]. In our cohort, we found that the expression of miR-101-3p and miR-423-5p in circulating exosomes and PBMCs was notably higher in MB patients than in healthy controls. A possible explanation for this may be that exosomes derived from monocytes/macrophages may act as a medium for the delivery of antitumorigenic factors. To further illustrate this issue, we isolated CD14+ monocytes directly from PBMC of plasma from MB patients and healthy controls, and detected the expression levels of miR-101-3p and miR-423-5p of the exosomes extracted from supernatants. The results showed that miR-101-3p and miR-423-5p were higher expressed in exosomes from the supernatant secreted by CD14+ monocytes in MB patients than in healthy controls. Together, the results illustrated the monocyte–macrophage is an origin of exosomes enriching miR-101-3p and miR-423-5p.
One limitation of our study was the relatively low sample number. We found that the levels of exosomal miR-101-3p and miR-423-5p were higher in plasma exosomes from MB patients than in those from healthy controls; however, no significant differences were observed between subgroups based on World Health Organization classification. A large population of MB patients will be required for further evaluation in the future.
In summary, we identified two exosomal miRNAs, miR-101-3p and miR-423-5p, as critical regulators of cell proliferation, migration, and apoptosis in MB, both in vitro and in vivo, and identified that these effects were exerted through direct binding to the FOXP4 3′UTR. We further found that miR-101-3p also targeted EZH2. Our data provide a novel therapeutic strategy for the treatment of MB (Fig. 8).
Materials and methods
Patients and ethics statement
Blood samples were obtained from MB patients and healthy donors at the Children’s Hospital of Fudan University, Shanghai, China, with written informed consent. Venous blood (4 ml) was collected in Vacutainer Plus blood collection tubes containing EDTA (BD Biosciences, San Jose, CA, USA). Blood plasma was retrieved from whole blood samples via centrifugation (300 × g, 10 min, 4 °C), aliquoted, and stored at −80 °C until analysis. The relevant characteristics of the patients are summarized in Supplementary Table 1. The study protocol was approved by the Ethics Committee of Children’s Hospital of Fudan University.
Exosome isolation and characterization
Exosomes were isolated from the plasma of MB patients, healthy control subjects, and culture supernatants of THP-1 and HMO6 cells by ultracentrifugation, as follows: 300 × g for 10 min, 2000 × g for 10 min to remove cell debris and large apoptotic bodies, followed by two rounds of ultracentrifugation at 100,000 × g for 70 min, with an intervening wash of pelleted exosomes with phosphate-buffered saline (PBS). Isolated exosomes were identified by morphological examination using a transmission electron microscope and western blotting-based detection of exosomal marker proteins. Exosomes were quantified using a BCA Protein Assay Kit (Takara Biomedical Technology, Beijing, China).
Nanoparticle tracking analysis
The particle size and concentration of isolated exosomes appropriately diluted in 1× PBS buffer were measured at VivaCellBiosceinces using the PMX 110 ZetaView NTA system (Particle Metrix, Meerbusch, Germany) and corresponding ZetaView 8.04.02 SP2 software. Measurements were recorded and analyzed at 11 positions. The ZetaView system was calibrated using 110-nm polystyrene particles. The temperature was maintained at approximately 23 and 30 °C.
Transmission electron microscopy
Purified exosome samples (10–20 µl) were placed on a carbon-coated copper grid for 1 min and then immersed in a 2% uranyl acetate solution for 1 min. Excess fluid was removed using filter paper and the exosomes imaged under a transmission electron microscope (JEM-1200EX; JEOL Ltd, Tokyo, Japan) at an accelerating voltage of 80 kV.
High-throughput sequencing of exosomal miRNA
Exosomal total RNA was isolated from plasma samples obtained from MB patients and healthy controls (n = 4 for each group). The genotypes of the four MB patients were one WNT type, two SHH types, and one Group 3 type. The preparation of miRNA sequencing libraries consisted of 3′-adaptor ligation, 5′-adaptor ligation, cDNA synthesis, PCR amplification, and size selection of PCR-amplified fragments of approximately 135–155 bps. The libraries were denatured to yield single-stranded DNA, captured on Illumina flow cells, amplified in situ as clusters, and finally sequenced for 50 cycles on the Illumina NextSeq platform.
After sequencing, Solexa CHASTITY quality-filtered reads were harvested as clean reads. The adaptor sequences were trimmed and miRDeep2 software was used to predict novel miRNAs from these adaptor-trimmed reads (≥15 nt). Then, the trimmed reads were aligned to merged pre-miRNA databases using NovoAlign software (v2.07.11) with a maximum of one mismatch permitted. Reads with counts <2 were discarded when calculating miRNA expression levels. To characterize isomiR variability, sequences that matched the miRNA precursors in the mature miRNA regions ± 4 nt (≤1 mismatch) were accepted as mature miRNA isomiRs and grouped according to the 5′ or 3′ arm of the precursor hairpin. The number of mapped tags of a specific miRNA was defined as its raw expression level. To correct for differences in tag counts among samples, tag counts were scaled to transcripts per million based on the total number of aligned tags. The most abundant isomiR, the mature miRNA annotated in miRBase, and all miRNA isoforms (5′ or 3′) were used to calculate miRNA expression levels. Fold-changes and p values were calculated and used to identify miRNAs significantly differentially expressed between groups. Hierarchical clustering was performed to identify distinguishable miRNA expression profiles among samples. MiRNA target prediction was performed using TargetScan and RNA22.
Exosome labeling and tracking
Purified exosomes isolated from plasma samples obtained from MB patients were labeled with PKH67 (Sigma-Aldrich, Darmstadt, Germany) following the manufacturer’s instructions. The exosomes were then re-extracted to remove free PKH67 dye using the SBI Exo-Quick TC kit (SBI, Beijing, China) and subsequently added to unstained Daoy cells to evaluate exosome uptake. Following incubation for 12 h at 37 °C, the cells were fixed, stained with Hoechst 33258 (1:4 000) for 10 min at room temperature, and then washed three times with PBS to remove excess dye. Cells were observed and imaged using a confocal microscope (Leica TCS-SP5 LSM, Wetzlar, Germany).
Cell culture and transfection
The human MB cell lines Daoy, and D283 Med, as well as the THP-1, HMO6, and HEK293T cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). All the cell lines have been validated using Short Tandem Repeat (STR) analysis and are kept mycoplasma-free. Daoy cells were maintained in α-MEM supplemented with 10% fetal bovine serum (FBS), 1% sodium pyruvate, 1% nonessential amino acid solution, and 1% penicillin/streptomycin (all from Gibco, Carlsbad, CA, USA). D283 Med cells and THP-1 monocytes were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin. HEK293T and HMO6 cells were maintained in high-glucose DMEM (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were transiently transfected with 200 nM chemically synthesized miR-101-3p and miR-423-5p mimics, siRNA targeting FOXP4 (siFOXP4) or EZH2 (siEZH2), or negative controls using Lipofectamine RNAiMAX Reagent (Invitrogen, Carlsbad, CA, USA). D283 Med cells were transfected with miR-101-3p/miR-423-5p lentiviral vectors or the negative control (GenePharma, Shanghai, China). The sequences of the mimics and siRNAs are listed in Supplementary Table 2.
Isolation of CD14+ monocytes and exosome collection
Peripheral blood mononuclear cells were isolated by Ficoll-Paque density gradient centrifugation (General Electric Healthcare, Piscataway, NJ, USA). CD14+ monocytes were further isolated using anti-human CD14 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions and cultured in RMPI 1640 medium supplemented with 10% exosome-free FBS and 1% penicillin/streptomycin for 72 h. Culture supernatants were harvested and exosomes were extracted for further miRNA detection.
RNA isolation and quantitative real-time PCR
Total RNA was extracted from cells and tissue using TRIzol reagent (Invitrogen). An ExoRNeasy Serum/Plasma Maxi Kit (Qiagen, Frankfurt, Germany) was used to extract exosomal total RNA. The expression levels of individual miRNAs were determined using miRNA sequence-specific probes (Thermo Fisher, Foster City, CA, USA). Beta-actin and U6 served as internal controls for mRNA and miRNA, respectively. Relative quantification was performed using the 2−ΔΔCT method. Primer sequences are listed in Supplementary Table 3.
Western blotting
Total protein was extracted and quantified, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, and transferred to a PVDF membrane (Millipore Corporation, Billerica, MA, USA), followed by blocking in 5% nonfat powdered milk at room temperature for 1 h. The membranes were incubated overnight at 4 °C with rabbit anti-CD9, anti-GM130, anti-EZH2, and anti-tubulin antibodies (13174, 12480, 5246; Cell Signaling Technology, Danvers, MA, USA), or anti-FOXP4 and anti-CD63 (ab17726, ab134045; Abcam, Cambridge, MA, USA). After being washed with 0.1% Tween-20-containing tris buffered saline solutions (TBST) for three times, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat-anti-rabbit antibody (Cell Signaling Technology) for 1 h. After being washed again with TBST for three times, protein bands were subsequently detected using a chemiluminescent HRP substrate (Thermo Fisher Scientific, Carlsbad, CA, USA) and imaged using a Bio-Rad ChemiDoc XRS + Imaging System.
CCK-8 and colony formation assays
Cell proliferation was evaluated by CCK-8 assay (Dojindo Molecular Technologies, Inc., Japan). Daoy cells were seeded into 96-well tissue culture plates at 1000 cells per well in complete medium and transfected with 200 nM miR-101-3p/miR-423-5p mimics, miR-NC, or siRNAs. D283 Med cells were transfected with the miR-101-3p/miR-423-5p lentiviral vectors or the negative control. The plates were incubated for a further 24 h, and then 10 µl of CCK-8 solution was added to each well, followed by incubation for an additional 1 h. The OD at 450 nm was measured at predetermined time points (0, 24, 48, or 72 h) on a Bio-Rad ELISA plate reader.
A colony formation assay was also performed to assess Daoy cell viability. Cells (500) were plated in each well of a six-well plate, transfected with the miR-101-3p/miR-423-5p mimics, siFOXP4, siEZH2, or negative control, and incubated at 37 °C for 2 weeks. The cells were fixed in methanol and then stained with 1% crystal violet. The number of cell colonies was determined and analyzed using ImageJ software.
Cell migration and invasion assays
The migratory ability of transfected Daoy cells was assessed by wound-healing and Transwell assays. For wound-healing assays, 1 × 105 cells were seeded into six-well plates until 90–100% confluence (48 h after transfection). A linear scratch wound was made across the middle of each well using the same pipette tip. The cells were then washed twice with PBS and incubated for 24 h with fresh culture medium without FBS. Images of the wound areas were captured using an inverted microscope.
For Transwell assays, 48 h after transfection, cells (2 × 104/100 µl) were plated into the upper chambers of 24-well Transwell plates in serum-free medium while MEM containing 10% FBS was added to the lower chambers. After incubation for 24 h at 37 °C, nonmigrated cells were removed with cotton swabs. Migrated cells were fixed in 4% paraformaldehyde for 20 min, stained with crystal violet solution for 20 min, and washed twice with PBS. Five visual fields were randomly chosen and imaged.
For the Transwell invasion assay, 2 × 104 cells in 100 µl of serum-free medium were added to the upper chambers of 24-well Transwell plates that had been pre-coated with Matrigel (100 µl/well) (BD Biosciences). The subsequent procedures were performed as described for the cell migration assays.
Apoptosis assay
Daoy cells (1 × 105) and D283 Med cells (1 × 105) were seeded into six-well plates. At 48 h after transfection, cells were treated with 100 μmol H2O2 overnight and then collected for apoptosis analysis. Apoptosis was evaluated by flow cytometry using a FITC–Annexin V Apoptosis Detection Kit I (BD, San Diego, CA, USA). After double staining with FITC-Annexin V/propidium iodide, the cells were analyzed using a FACS Canto II Cell Analyzer (BD Biosciences). The results were analyzed using FlowJo software (Tree Star Inc., San Jose, CA, USA). The percentages of apoptotic cells were calculated as the sum of the percentages from quadrant 3 (early apoptosis) and quadrant 2 (late apoptosis).
Luciferase reporter assays
HEK293T cells were co-transfected with the psiCHECK-2 plasmid containing the wild type or mutated FOXP4 or EZH2 3′UTR (Promega, Fitchburg, WI, USA) and miR-101-3p/miR-423-5p mimics or respective controls using Lipofectamine 2000 (Invitrogen) in 96-well plates. Firefly and Renilla luciferase activities were measured 24 h after transfection using a dual-luciferase reporter assay kit (Promega) following the manufacturer’s protocol. Renilla luciferase activity values were used for normalization.
In vivo experiments
The lentiviral vectors for the delivery of miR-101-3p and miR-423-5p and the corresponding negative controls were purchased from GenePharma. Athymic male BALB/c nude mice, 5–6 weeks old, were maintained under specific pathogen-free conditions in the Experimental Animal Department of Fudan University. Mice were randomly assigned to one of three groups (n = 6 mice per group). Daoy cells (5 × 106) transfected with LV16-miR-101-3p, LV16-miR-423-5p, or LV16-NC were subcutaneously injected into the right flank of each mouse in a volume of 100 µl mixed with 30% Matrigel. When palpable tumors had developed, caliper measurements were taken once a week. Tumor volumes were calculated using the following formula: volume = 0.5 × length × width2. A tumor growth curve was generated based on tumor volume and the corresponding treatment duration. Mice were sacrificed 7 weeks postinjection and xenograft-derived tumors were excised and weighed. All experimental protocols were approved by the Animal Studies Committee of the Children’s Hospital of Fudan University. Experiments using animals were performed in duplicate.
Immunohistochemical staining
Tumor tissues were fixed and embedded in paraffin before deparaffinization and rehydration for antigen retrieval. Sections were incubated with primary antibodies against Ki-67(BD Biosciences), EZH2 (Cell Signaling Technology), and FOXP4 (Abcam) at 4 °C overnight, followed by incubation with secondary biotinylated antibodies. The sections were visualized with DAB solution and counterstained with hematoxylin. Images were captured under a light microscope (Leica).
Statistical analysis
Data are presented as means ± SEM from three independent experiments. Statistical analysis was performed using the Student’s t-test for between-group comparisons or one-way ANOVA for multiple group comparisons in GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA). Differences were considered significant when p < 0.05.
Data availability
The sequencing data have been deposited in the Gene Expression Omnibus (GEO) under accession numbers GSE123376.
References
Kohler BA, Ward E, McCarthy BJ, Schymura MJ, Ries LA, Eheman C, et al. Annual report to the nation on the status of cancer, 1975-2007, featuring tumors of the brain and other nervous system. J Natl Cancer Inst. 2011;103:714–36.
Massimino M, Biassoni V, Gandola L, Garre ML, Gatta G, Giangaspero F. et al. Childhood medulloblastoma. Crit Rev Oncol Hematol. 2016;105:35–51.
Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta Gen. Subj. 2012;1820:940–8.
Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–4.
Leichter AL, Sullivan MJ, Eccles MR, Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol Cancer. 2017;16:15.
Bell E, Taylor MA. Functional roles for exosomal microRNAs in the tumour microenvironment. Comput Struct Biotechnol J. 2017;15:8–13.
Sidhom K, Obi PO, Saleem A. A review of exosomal isolation methods: is size exclusion chromatography the best option?. Int J Mol Sci. 2020;21:6466
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54.
Liu Y, Luo F, Wang BR, Li HQ, Xu Y, Liu XL. et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370:125–35.
Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS. et al. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–52.
Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46.
Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21.
Samsonov R, Shtam T, Burdakov V, Glotov A, Tsyrlina E, Berstein L. et al. Lectin-induced agglutination method of urinary exosomes isolation followed by mi-RNA analysis: application for prostate cancer diagnostic. Prostate. 2016;76:68–79.
Fong MY, Zhou WY, Liu L, Alontaga AY, Chandra M, Ashby J. et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183–94.
Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. Delivery of functional Anti-miR-9 by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol Ther Nucl Acids. 2013;2:e126.
Song L, Huang Q, Chen K, Liu L, Lin C, Dai T. et al. miR-218 inhibits the invasive ability of glioma cells by direct downregulation of IKK-beta. Biochem Biophys Res Commun. 2010;402:135–40.
Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, et al. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci USA. 2009;106:2812–7.
Huang S, Xue P, Han X, Zhang C, Yang L, Liu L. et al. Exosomal miR-130b-3p targets SIK1 to inhibit medulloblastoma tumorigenesis. Cell Death Dis. 2020;11:408
Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K. et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121:984–95.
Teufel A, Wong EA, Mukhopadhyay M, Malik N, Westphal H. FoxP4, a novel forkhead transcription factor. Biochim Biophys Acta Gene Struct Expr. 2003;1627:147–52.
Tsang DPF, Cheng ASL. Epigenetic regulation of signaling pathways in cancer: Role of the histone methyltransferase EZH2. J Gastroenterol Hepatol. 2011;26:19–27.
McCabe MT, Creasy CL. EZH2 as a potential target in cancer therapy. Epigenomics. 2014;6:341–51.
Alimova I, Venkataraman S, Harris P, Marquez VE, Northcott PA, Dubuc A, et al. Targeting the enhancer of zeste homologue 2 in medulloblastoma. Int J Cancer. 2012;131:1800–9.
Jia WZ, Yu T, An Q, Cao XL, Pan HD. MicroRNA-423-5p inhibits colon cancer growth by promoting caspase-dependent apoptosis. Exp Ther Med. 2018;16:1225–31.
Tang XB, Zeng XG, Huang Y, Chen SB, Lin F, Yang G. et al. miR-423-5p serves as a diagnostic indicator and inhibits the proliferation and invasion of ovarian cancer. Exp Ther Med. 2018;15:4723–30.
Li J, Liu X, Fang Q, Ding M, Li C. Liraglutide attenuates atherosclerosis via inhibiting ER-induced macrophage derived microvesicles production in T2DM rats. Diabetol Metab Syndr. 2017;9:94.
Yang H, Fu HL, Wang B, Zhang X, Mao JH, Li X. et al. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol Carcinog. 2018;57:1223–36.
Yao YL, Ma J, Wang P, Xue YX, Li Z, Zhao LN. et al. miR-101 acts as a tumor suppressor by targeting Kruppel-like Factor 6 in glioblastoma stem cells. CNS Neurosci Ther. 2015;21:40–51.
Yang T, Li H, Thakur A, Chen TJ, Xue J, Li D. et al. FOXP4 modulates tumor growth and independently associates with miR-138 in non-small cell lung cancer cells. Tumor Biol. 2015;36:8185–91.
Wang G, Sun YB, He YF, Ji CS, Hu B, Sun YP. MicroRNA-338-3p inhibits cell proliferation in hepatocellular carcinoma by target forkhead box P4 (FOXP4). Int J Clin Exp Pathol. 2015;8:337–44.
Yin ZX, Ding HM, He EX, Chen JC, Li M. Up-regulation of microRNA-491-5p suppresses cell proliferation and promotes apoptosis by targeting FOXP4 in human osteosarcoma. Cell Prolif. 2017;50:e12308.
Zingg D, Debbache J, Pena-Hernandez R, Antunes AT, Schaefer SM, Cheng PF, et al. EZH2-mediated primary cilium deconstruction drives metastatic melanoma formation. Cancer Cell. 2018;34:69–84.
Oki S, Sone K, Oda K, Hamamoto R, Ikemura M, Maeda D. et al. Oncogenic histone methyltransferase EZH2: a novel prognostic marker with therapeutic potential in endometrial cancer. Oncotarget. 2017;8:40402–11.
Labbe DP, Sweeney CJ, Brown M, Galbo P, Rosario S, Wadosky KM. et al. TOP2A and EZH2 provide early detection of an aggressive prostate cancer subgroup. Clin Cancer Res. 2017;23:7072–83.
Hirukawa A, Smith HW, Zuo DM, Dufour CR, Savage P, Bertos N. et al. Targeting EZH2 reactivates a breast cancer subtype-specific anti-metastatic transcriptional program. Nat Commun. 2018;9:2547.
Sanna L, Marchesi I, Melone MAB, Bagella L. The role of enhancer of zeste homolog 2: from viral epigenetics to the carcinogenesis of hepatocellular carcinoma. J Cell Physiol. 2018;233:6508–17.
Su MJ, Aldawsari H, Amiji M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of microRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci Rep. 2016;6:30110.
Zhang C, Wang H, Liu X, Hu Y, Ding L, Zhang X. et al. Oncogenic microRNA-411 promotes lung carcinogenesis by directly targeting suppressor genes SPRY4 and TXNIP. Oncogene. 2019;38:1892–904.
Xiao M, Zhang J, Chen W, Chen W. M1-like tumor-associated macrophages activated by exosome-transferred THBS1 promote malignant migration in oral squamous cell carcinoma. J Exp Clin Cancer Res. 2018;37:143
Acknowledgements
We acknowledge and appreciate the colleagues from our lab for their valuable efforts and comments on this paper.
Funding
This work was supported by grants from the National Key R&D Program of China (2016YFC1305102 to YZ), National Natural Science Foundation of China (81671561, 81974248 to YZ, 81900751 to XH), International Joint Laboratory Program of the National Children’s Medical Center (EK1125180109 to YZ), Program for Outstanding Medical Academic Leader (2019LJ19 to YZ), and Shanghai Committee of Science and Technology (21140902400 to YZ, 21ZR1410000 and 19ZR1406400 to JF).
Author information
Authors and Affiliations
Contributions
These authors contributed equally: PX, SH, and XH. PX, SH, and XH designed and performed experiments and analyzed data. CZ and LY generated mice model together with PX and SH. JF and WX designed experiments and analyzed data. PX and SH analyzed data, prepared figures, and wrote the manuscript. HL recruited the human participants. YZ planned, designed, supervised, and coordinated the overall research efforts.
Corresponding authors
Ethics declarations
competing interests
The authors declare no competing interests.
Ethical approval
The study protocol was approved by the Ethics Committee of Children’s Hospital of Fudan University. Informed consent was obtained from all participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Piacentini
Rights and permissions
About this article
Cite this article
Xue, P., Huang, S., Han, X. et al. Exosomal miR-101-3p and miR-423-5p inhibit medulloblastoma tumorigenesis through targeting FOXP4 and EZH2. Cell Death Differ 29, 82–95 (2022). https://doi.org/10.1038/s41418-021-00838-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00838-4
This article is cited by
-
miR-101-3p-mediated role of PDZK1 in hepatocellular carcinoma progression and the underlying PI3K/Akt signaling mechanism
Cell Division (2024)
-
miR-124-3p and miR-194-5p regulation of the PI3K/AKT pathway via ROR2 in medulloblastoma progression
Cancer Gene Therapy (2024)
-
Analysis on EZH2: mechanism identification of related CeRNA and its immunoassay in hepatocellular carcinoma
BMC Medical Genomics (2023)
-
miR-423 sponged by lncRNA NORHA inhibits granulosa cell apoptosis
Journal of Animal Science and Biotechnology (2023)
-
Exosomes in the tumor microenvironment of cholangiocarcinoma: current status and future perspectives
Journal of Translational Medicine (2022)