Abstract
The human oral and gut commensal microbes play vital roles in the development and maintenance of immune homeostasis, while its association with susceptibility and severity of SARS-CoV-2 infection is barely understood. In this study, we investigated the dynamics of the oral and intestinal flora before and after the clearance of SARS-CoV-2 in 53 COVID-19 patients, and then examined their microbiome alterations in comparison to 76 healthy individuals. A total of 140 throat swab samples and 81 fecal samples from these COVID-19 patients during hospitalization, and 44 throat swab samples and 32 fecal samples from sex and age-matched healthy individuals were collected and then subjected to 16S rRNA sequencing and viral load inspection. We found that SARS-CoV-2 infection was associated with alterations of the microbiome community in patients as indicated by both alpha and beta diversity indexes. Several bacterial taxa were identified related to SARS-CoV-2 infection, wherein elevated Granulicatella and Rothia mucilaginosa were found in both oral and gut microbiome. The SARS-CoV-2 viral load in those samples was also calculated to identify potential dynamics between COVID-19 and the microbiome. These findings provide a meaningful baseline for microbes in the digestive tract of COVID-19 patients and will shed light on new dimensions for disease pathophysiology, potential microbial biomarkers, and treatment strategies for COVID-19.
Similar content being viewed by others
Introduction
The ongoing COVID-19 pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has jeopardized global public health and safety. As of June 21, 2021, the COVID-19 pandemic has infected more than 179 million people, resulting in over 3.8 million causalities globally. Transmission of SARS-CoV-2 has commonly occurred via breathing or direct contact with virus-containing droplets and aerosols from infected people through coughing and sneezing. When the SARS-CoV-2 particle reaches the nasal cavity of the host, it enters the host epithelial cells through the angiotensin-converting enzyme 2 (ACE2) receptor, which is prominently presented on epithelial cells lining in the respiratory and digestive tract systems1,2. Most infections lead to a prompt innate immune response and the virus gets eradicated or controlled quickly, manifesting none or mild symptoms. Whereas in some patients, viruses located in the upper airway replicate further toward the lower respiratory tract to activate enhanced pro-inflammatory responses, therefore resulting in severe outcomes including acute respiratory syndrome, organ malfunctioning, and even death3. Not limited to the respiratory tract, SARS-CoV-2 is known to target different organ systems4. For instance, around 55% of patients were observed with prolonged viral RNA present in the feces, even weeks after the viral clearance in their respiratory tract5. It has been reported that abnormal immune response, comorbid conditions, and advanced age are risk factors linked to COVID-19 severity. However, these factors are insufficient to offer a satisfactory explanation of all patients’ severe disease outcomes. As global mass vaccination and proven effective therapy for COVID-19 remain unclear, explorative efforts towards new perspectives of protection and therapeutic approaches for COVID-19 are indispensable6,7.
The human microbiome is important for developing and maintaining immune homeostasis and it is known that microbiota imbalance or dysbiosis are highly associated with various diseases. The intestinal tract and oral cavity, with the largest and second-largest microbiota in the human body, play significant roles in the pathogenesis of infectious disease. Previous studies have reported that oral-lung microbes can influence the outcome of many infectious diseases by regulating the host mucosal immunity8,9,10. Intestinal flora can affect the occurrence and progression of viral infection through the gut–lung axis11,12,13. Conversely, the indigenous microbiome could be disturbed by a viral infection, leading to alterations in susceptibility and disease severity through dysbiotic community structure and function14. The unequivocal association between influenza viral and bacterial co-infection and disease severity has been proved in early studies15. Likewise, evidence has suggested that SARS-CoV-2 infection could predispose patients to bacterial co-infections and superinfections, resulting in increased disease severity and mortality14. Further, the dysbiosis of influenza-infected individuals progresses toward microbiota homeostasis, coinciding with viral clearance and recovery, suggesting that the health status of microbial flora is likely a fine indication of disease recovery16. Zuo and colleagues have also suggested that fecal microbiomes from COVID-19 patients were characterized by proliferation of opportunistic pathogens and depletion of favorable commensals compared to healthy controls with the shot-gun metagenomics approach17; however, this study was limited by its size of subjects used. Moreover, validation of the profiles at the genus level is currently suggested to be performed through 16S rRNA gene microbial profiling to evaluate the presence of undetected microbes in the marker gene-based profiles18. Accumulated evidence for the oral–gut axis has revealed its role in modulating the pathogenesis process in numerous diseases19,20. It is compelling to look into the oral and intestinal microbiome combined with SARS-CoV-2 infection and the crosstalk among them, which may provide an improved understanding of the initiation of viral infection and the path of disease deterioration.
Results
Overview of microbial composition in the study subjects
A total of 53 patients diagnosed with COVID-19 and 76 healthy individuals were included in this study. As indicated in Fig. 1a, serial throat swab samples and fecal specimens were collected from COVID-19 patients during hospitalization, covering both the positive viral RNA test period (P-VRTP) and the negative viral RNA test period (N-VRTP). Depending on the sample availability, a total of 140 throat swab samples, including 52 during the P-VRTP [PT(+)] and 88 during the N-VRTP [PT(−)], and a total of 81 fecal samples, including 50 during the P-VRTP [PF(+)] and 31 during the N-VRTP [PF(−)] were collected from hospitalized patients (P: patient). In addition, 44 throat swab samples (HT) and 32 fecal samples (HF) from sex and age-matched healthy individuals (H: healthy individual) were included in our study as controls. The sample distribution, demographics, and relevant clinical information about the subjects recruited in the study were summarized in Supplementary Table 1.
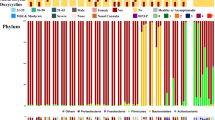
a Graphic representation of study design and sample collection. PT(+), throat swab samples from COVID-19 patients during the positive viral RNA test period; PT(−), throat swab samples from COVID-19 patients during the negative viral RNA test period; PF(+), fecal specimens from COVID-19 patients during the positive viral RNA test period; PF(−), fecal specimens from COVID-19 patients during the negative viral RNA test period. b Bacterial composition at the phylum level among groups. Sample size (n) for each group: HT, n = 44; PT(+), n = 52; PT(−), n = 88; HF, n = 32; PF(+), n = 50; PF(−), n = 31. HT, throat swab samples from the healthy controls; HF, fecal specimens from the healthy controls. c PCoA plot based on the unweighted UniFrac distance depicting differences in the bacterial community among groups.
To characterize the microbiotas in those aforementioned samples, we sequenced the V3–V4 region of the bacterial 16S rRNA gene. As expected, our data confirmed a distinct microbiota composition between oral and gut samples21. The three most abundant phyla Firmicutes, Proteobacteria, and Bacteroidetes accounted for 79.3% of the community in the oral samples. The two dominant phyla Firmicutes and Bacteroidetes accounted for 83.7% of the bacterial community in the gut samples (Fig. 1b). The difference was further illustrated by the principal coordinates analysis (PCoA) plot and Permutational multivariate analysis of variance (PERMANOVA) with unweighted UniFrac distance calculated at the sequence feature level. The microbiota composition of the oral and gut samples diverged from each other along the first axis in which 31.87% of the total variance was explained, indicating that the largest source of variation was the sample types (Fig. 1c). Similarly, the PERMANOVA result indicated 30.80% of the total variance was explained by the sample types (oral samples vs. gut samples, PERMANOVA, R2 = 0.308, p < 0.001). More importantly, samples from patients and healthy individuals seemed to possess strong dissimilarities for both oral (PERMANOVA, R2 = 0.175, p < 0.001) and gut microbiome communities (PERMANOVA, R2 = 0.196, p < 0.001), suggesting overwhelming shifts of microbiome structure were gained after SARS-CoV-2 infection in both oral cavity and feces. Meanwhile, the microbial compositions of COVID-19 patients collected during the P-VRTP and the N-VRTP presented no significant discrepancy in oral samples (PERMANOVA, R2 = 0.012, p = 0.080) or in gut samples (PERMANOVA, R2 = 0.016, p = 0.139). Due to their distinctive characteristics, the oral and gut microbiome profiles and their potential roles in SARS-CoV-2 infection were addressed in separate sections followed.
Alterations of the oral microbiota in patients
Given the observation that the SARS-CoV-2 infection may impact the structure of the microbiome (Fig. 1c), we first investigated the effects of different disease severity conditions and treatment therapies on the oral samples from patients. The diversity metrics among oral microbiomes from the healthy controls (HT), patients with the non-severe condition [PT(NS)], and patients with the severe condition [PT(S)] during hospitalization were compared. Alpha diversity of Faith’s phylogenetic index revealed that the oral microbial diversity was decreased in COVID-19 patients compared to the healthy controls (ANOVA, PT(NS) vs. HT, p = 0.016; PT(S) vs. HT, p < 0.0001). Further, a significant deduction of diversity was observed in the PT(S) group when comparing to the PT(NS) group (ANOVA, p < 0.0001, Fig. 2a). Similarly, inter-individual beta diversity unweighted UniFrac dissimilarity revealed that the microbiomes of the PT(NS) group (PERMANOVA, R2 = 0.238, p < 0.001) and the PT(S) group (PERMANOVA, R2 = 0.233, p < 0.001) clustered apart from that of the healthy controls. We also found a significant dissimilarity between the PT(S) group and the PT(NS) group (PERMANOVA, R2 = 0.049, p < 0.001, Fig. 2b). The significant differences were partially attributed to the different dispersions between the PT(S) group and the PT(NS) group (PERMDISP, p = 0.002). In contrast, the difference in dispersion between the HT group and the PT(NS) group was not significant (PERMDISP, p = 0.166). We also investigated the impact of clinical antibiotic usage. The PCoA based on unweighted UniFrac dissimilarity demonstrated that bacterial communities in the HT group, the patients without antibiotic interference [PT(abx−)] group and the patients with antibiotic interference [PT(abx+)] group were mutually separated (PERMANOVA, PT(abx−) vs. HT, R2 = 0.258, p = 0.001; PT(abx+) vs. HT, R2 = 0.204, p = 0.001; PT(abx−) vs. PT(abx+), R2 = 0.016, p = 0.016; Fig. 2c). To identify potential microbial biomarkers associated with COVID-19 patients, we employed a combined approach with LEfSe and MaAsLin2 to minimize the influence of confounding factors. Seventeen significantly different taxa were identified in the oral microbiome between the healthy controls and COVID-19 patients (HT and PT, Fig. 2d and Supplementary Table 2). Specifically, significant decreases of Neisseria, Corynebacterium, Actinobacillus, Moryella, Aggregatibacter, Treponema, and Pseudomonas at the genus level, as well as P. intermedia and T. amylovorum were observed in the patients comparing to the controls. In contrast, Veillonella, Campylobacter, Granulicatella, Kingella, and Filifactor at the genus level, as well as H. parainfluenzae, R. mucilaginosa, and N. subflava were enriched in the oral microbial communities of COVID-19 patients compared to those of the controls. A similar comparison was conducted between the PT(NS) and the PT(S) groups to assess the impact of disease severity on bacteria (Supplementary Table 3). Bacterial taxa including Treponema, Aggregatibacter, and P. intermedia were identified further depleted in the PT(S) group, implying their associations with disease severity. The comparison between the controls and the PT(abx–) group was carried out to assess the impact of infection on bacteria by excluding the antibiotics interference (Supplementary Table 3). Lastly, the potential functional consequence resulted from the drastic taxa compositional alteration was evaluated. The global pattern from Procrustes analysis displayed a good-fit correlation between the oral microbial community composition and microbial function (Mantel, rho = 0.403, p = 0.001, Supplementary Fig. 1a). The significant difference in functional pathways of the oral microbial communities between COVID-19 patients and the controls was shown in Fig. 2e. Notably, the top four depleted pathways in patients were involved in the TCA cycle used by all aerobic organisms to generate energy, indicating a disturbed microbial community.
a Species diversity differences among the HT (n = 44), PT(NS) (n = 90), and PT(S) (n = 50) groups were estimated by Faith’s phylogenetic diversity index. *p < 0.05, ****p < 0.0001. p-values were obtained using one-way ANOVA followed by Tukey’s multiple comparisons test. The line in the middle of the box, bound of the box and whiskers represent the median, 25th–75th percentiles, and min-to-max values, respectively. PT(NS), throat swab samples from the non-severe patients group; PT(S), throat swab samples from the severe patients group. b PCoA plot based on unweighted UniFrac distances showing microbial structural clustering among the HT, PT(NS), and PT(S) groups. Ellipses represent 68% confidence intervals. c PCoA plot according to unweighted UniFrac distances displaying bacterial structural discrimination among the HT (n = 44), PT(abx−) (n = 49), and PT(abx+) (n = 91) groups. Ellipses represent 68% confidence intervals. PT(abx−), throat swab samples from patients without antibiotic interference; PT(abx+), throat swab samples from patients with antibiotic interference. d Bacterial taxa identified a significant difference between HT (n = 44) and PT (n = 140) groups by LEfSe. Seventeen taxa were validated using MaAsLin2 adjusting for covariates. PT throat swab samples from patients, LDA linear discriminant analysis. e Differentially abundant MetaCyc pathways identified with the functional analysis result of metagenomes. Only the pathways identified by both LEfSe and MaAsLin2 were presented. Circle size represents the relative abundance of pathways.
Alterations of the gut microbiota in patients
Next, we explored whether dysbiosis occurred in the gut microbiome. Faith’s phylogenetic diversity index showed that its diversity was significantly lower in patients with the non-severe condition [PF(NS)] (ANOVA, p < 0.0001) and patients with the severe condition [PF(S)] (ANOVA, p < 0.0001) in comparisons to the healthy controls (HF, Fig. 3a). The diversity of the PF(NS) group was slightly higher than the PF(S) group (ANOVA, p = 0.73). Besides, the PCoA plot with unweighted UniFrac distances revealed a cluster separation in the fecal microbiota between PF(NS) and PF(S) (PERMANOVA, R2 = 0.067, p < 0.001), and between the patients and the healthy controls (PERMANOVA, PF(NS) vs. HF, R2 = 0.232, p < 0.001; PF(S) vs. HF, R2 = 0.267, p < 0.001, Fig. 3b). Unlike the oral microbiome, the gut microbiota in the PF(NS) group seemed more impacted after infection, as reflected by its greater statistical dispersion than the HF group (PERMDISP, p = 0.002). Moreover, the gut microbiota among patients with antibiotic interference [PF(abx+)], patients without antibiotic interference [PF(abx−)], and the HF group exhibited mutual clustering separation with unweighted UniFrac distances based PCoA plot (PERMANOVA, PF(abx−) vs. HF, R2 = 0.262, p = 0.001; PF(abx+) vs. HF, R2 = 0.230, p = 0.001; PF(abx−) vs. PF(abx+), R2 = 0.030, p = 0.002; Fig. 3c). In terms of potential microbial biomarkers, we identified 17 bacterial taxa again with confounding covariates adjusted. Concretely, decreased taxa included Blautia, Coprococcus, and Collinsella at the genus level, and B. caccae, B. coprophilus, B. obeum, and C. colinum species; and increased taxa included Streptococcus, Weissella, Enterococcus, Rothia, Lactobacillus, Actinomyces, and Granulicatella at the genus level as well as C. citroniae, B. longum, and R. mucilaginosa species (Fig. 3d and Supplementary Table 4). More interestingly, two normal components of the respiratory tract flora, Granulicatella and R. mucilaginosa, were significantly increased in the patients for both oral and gut samples, and R. mucilaginosa seemed further associated with the disease severity in the fecal samples (Supplementary Fig. 2). Additional analysis focusing on the bacteria changes associated with disease severity and antibiotics usage in fecal samples were described in Supplementary Table 5. Notably, no difference in the bacterial taxa was identified between the PF(NS) group and the PF(S) group. Similar to the oral microbiome, the global pattern from Procrustes analysis displayed a good-fit correlation between the microbial composition and predicted function profile in the gut microbiota community (Mantel, rho = 0.268, p = 0.001, Supplementary Fig. 1b). Also, the major distinct functional pathways associated with SARS-CoV-2 infection were shown in Fig. 3e, including two depleted pyrimidine deoxyribonucleotides biosynthesis pathways involved in viral suppression through innate immunity in patients22.
a Species diversity differences among the HF (n = 32), PF(NS) (n = 64), and PF(S) (n = 17) groups were estimated by Faith’s phylogenetic diversity index. ns: not significant, *p < 0.05, ****p < 0.0001. p-values were obtained using one-way ANOVA followed by Tukey’s multiple comparisons test. The line in the middle of the box, bound of the box and whiskers represent the median, 25th–75th percentiles, and min-to-max values, respectively. PF(NS), fecal specimens from the non-severe patient group; PF(S), fecal specimens from the severe patient group. b PCoA plot based on unweighted UniFrac distances showing microbial structural clustering among the HF, PF(NS), and PF(S) groups. Ellipses represent 68% confidence intervals. c PCoA plot according to unweighted UniFrac distances displaying bacterial structural discrimination among the HF (n = 32), PF(abx−) (n = 30), and PF(abx+) (n = 51) groups. Ellipses represent 68% confidence intervals. PF(abx−), fecal specimens from patients without antibiotic interference; PF(abx+), fecal specimens from patients with antibiotic interference. d Bacterial taxa identified a significant difference between the HF (n = 32) and PF (n = 81) groups by LEfSe. Seventeen taxa were validated using MaAsLin adjusting for covariates. PF fecal samples from patients, LDA linear discriminant analysis. e Differentially abundant MetaCyc pathways identified with the functional analysis result of metagenomes. Only the pathways identified by both LEfSe and MaAsLin2 were presented. Circle size represents the relative abundance of pathways.
Relationship between SARS-CoV-2 virus and bacterial species
The viral loads in the throat swab and fecal samples spanning the full collection period were compared. The median time span of the throat swabs collection was 10 (IQR: 6–12) days for P-VRTP and 10 (IQR: 3–18) days for N-VRTP. The median time span of the fecal samples collection was 8 (IQR: 3.75–14) days for P-VRTP and 8 (IQR: 2–12) days for N-VRTP. The viral copy numbers derived from the E gene showed a relatively higher abundance over the numbers derived from the N gene (Fig. 4a). As E-gene and N-gene copy numbers showed a strong correlation therefore both can accurately reflect the abundance of viral copy (throat swab samples: Spearman rho = 0.872, p < 0.001; fecal samples: Spearman rho = 0.923, p < 0.001). The overall viral loads in throat swab samples seemed equally abundant with fecal specimens collected from patients during the P-VRTP, which is consistent with the previous studies23. Based on unweighted UniFrac distance, it was found that the differences in beta diversity between the HT and PT(+) groups (PERMANOVA, R2 = 0.234, p = 0.001), the HT and PT(−) groups (PERMANOVA, R2 = 0.218, p = 0.001), were significant from each other. Likewise, unweighted UniFrac dissimilarity revealed that the microbiomes of the PF(+) group (PERMANOVA, R2 = 0.233, p = 0.001) and the PF(−) group (PERMANOVA, R2 = 0.250, p = 0.001) were significantly different from that of the healthy controls. During the N-VRTP, we observed a microbial community recovery towards the healthy controls, that is, unweighted UniFrac distance between PT(−) vs. HT was lower than that between PT(+) vs. HT (Fig. 4b). Compared to the oral microbiome, this recovery trend in the microbiome in the gut was less evident, which may be attributed to a relatively shorter time span of sample collection and microbiome robustness in the fecal samples. Additionally, we investigated whether the SARS-CoV-2 viral load was associated with any oral and gut bacterial species. In the oral microbiota, Pelomonas was identified as positively correlated with the viral load of SARS-CoV-2 (Fig. 4c). While in the gut microbiota, P. copri and E. dolichum were identified positively correlated with the viral load of SARS-CoV-2, and S. anginosus, Dialister, Alistipes, Ruminococcus, C. citroniae, Bifidobacterium, Haemophilus, and H. parainfluenzae were identified negatively correlated with the viral load of SARS-CoV-2.
a Longitudinal changes of viral loads in throat swab samples and fecal specimens from COVID-19 patients. Day 0 indicated the negative conversion time of SARS-CoV-2 RNA. The red line and blue line indicated longitudinal changes in viral loads represented by the abundances of the E gene and N gene, respectively. b Boxplots showing unweighted UniFrac distances within and between groups. c The longitudinal viral load represented by the abundances of the E gene significantly correlated with the timepoint-matched relative abundances of different taxa in throat swab and fecal samples separately. p-values were obtained using CCLasso.
Discussion
The oral and gut microbiome offers a range of valuable properties to the host. Several most significant contributions of these microbes are to boost metabolism, improve digestive health and strengthen resistance against pathogens. Furthermore, the complex crosstalk between commensal microbes and different body systems is essential for the functioning of the immune system24.
Our data revealed profound alterations in both oral and gut microbiomes, which were reflected in the dramatic changes in community structure, potential bacterial marker species, and predicted functional profile. The decline in commensal bacterial diversity has been considered as a key dysbacteriosis indicator in several diseases25. Coherently, the oral and gut microbiome of COVID-19 patients in our study exhibited decreased diversities compared to the healthy controls. This trend of microbial imbalance in patients with severe conditions compared to non-severe patients was also observed in both sample types, though the alpha diversity was not significant (ANOVA, p = 0.73) in the fecal samples. Moreover, the microbiomes from the severe and the non-severe patients were partitioned into two clusters in both microbial populations. Overall, diversity results of impaired microbiota suggested the strong association between the microbiome community complexity and the disease severity in COVID-19 patients. Though experimental validation is needed, our result has highlighted the possibility of personalized microbiota to affect the disease outcome of COVID-19 patients26.
Numerous studies have reported high occurrences of bacterial co-infection in hospitalized COVID-19 patients, and the odds of the bacterial infection get even higher for ICU patients27,28. Among the list of eight bacterial taxa with enlarged relative abundance in patients in Fig. 2d, Veillonella, Campylobacter, R. mucilaginosa, Granulicatella, Kingella, and Filifactor belongs to a group of periodontitis-correlated taxa, adding evidence to support the previous work denotative of a close relationship between periodontitis and SARS-CoV-2 infection29. Periodontal-associated cytokines may proliferate contacts of bacteria between the lungs and the mouth via driving the alteration of the respiratory epithelium and thereby promote respiratory infection in COVID-19 patients14. H. parainfluenzae and Kingella, two enriched taxa in the oral cavity of COVID-19 patients, are opportunistic pathogens well-known for respiratory tract infections, infective endocarditis, and meningitis30,31. The abrupt loss of Neisseria in patients, which is the highly abundant genus in the normal oral cavity, could raise serious damage to the oral microbiota32,33. In addition, all increased bacteria were previously classified as bacteremia-associated bacteria, implying a potential association between oral dysbiosis and secondary bacterial infection in COVID-19 patients34. Typically, mechanical ventilation supports may predispose the COVID-19 patients with severe symptoms of dyspnea to pulmonary bacterial co-infection, as the penetration of the device provides an entrance for those opportunistic infectious agents to access the lower respiratory tract from the oral cavity. Given the non-ignorable association between the oral microbiome with bacterial co-infections suggested by our and others’ data35,36, correct and frequent oral health care measurements should be recommended by physicians to protect COVID-19 patients from secondary infections and improve survival, especially for the ones with the severe disease condition.
Elevated levels of Streptococcus, Rothia, and Actinomyces were identified in COVID-19 patients’ feces, which is consistent with previous findings37. The host immunity symbionts beneficial bacterial species B. obeum, whose parent genus Blautia was listed as the top decreased taxon in COVID-19 patients’ feces in our data, was identified to be depleted in another study17. Notably, two normal components of the respiratory tract flora, Granulicatella, and R. mucilaginosa, were identified enriched in patients’ feces, which is likely a reflection of the migration of extra-intestinal microbes into the gut or the flourishment of potentially pathogenic bacteria. Typically, the fecal enrichment of specific oral taxa, which was suggested to be linked with the increased oral–fecal microbial transmission, has frequently been regarded as a hallmark of disease20. As SARS-CoV-2 viral RNA was detected in stools, the route of viral transmission from the respiratory tract to the intestinal tract can be hinted by the path of the oral flora translocation and colonization38. Moreover, R. mucilaginosa seemed further associated with the disease severity in the fecal samples in our study. As some severe patients with COVID-19 linked with cardiovascular disease comorbidities, this finding supports previous work wherein patients with cardiovascular disease comorbidities tended to have a higher prevalence of the Rothia ASV associated with SARS-CoV-239,40. In addition, increased expression of ACE2 receptor was observed in B. longum treated mice; hence changes of B. longum might impact the individual’s susceptibility to SARS-CoV-2 through modulating ACE2 expression41. It is worth noticing that a decreased relative abundance of B. caccae and B. coprophilus was observed in the gut flora of COVID-19 patients. A recent study by Martino et al.42, has demonstrated that Bacteroides may regulate viral adhesion via modifying heparan sulfate (HS), and loss of HS-modifying Bacteroides strains could predispose individuals to SARS-CoV-2 infection. Concordance of these findings with ours provides evidence for a potential relationship between B. caccae and B. coprophilus and susceptibility to SARS-CoV-2 infection.
The Mantel and Procrustes analysis indicated the predicted function profiles aligned well with the bacterial taxonomy profile in both oral and fecal samples as expected. Compared to the gut microbiome, our analysis suggested a stronger impairment in the oral microbiome after SARS-CoV-2 infection, wherein a significant reduction of Neisseria, an essential oral microbial genus, was found, along with several fatal metabolic pathways involving the TCA cycle were suppressed. The result echoes a previous study that the gut microbial community possesses higher taxa-function robustness over oral community43, since the bacterial community in the gut generally has a higher gene and functional redundancy in comparison to communities inhabiting other body sites.
Microbiome dysbiosis persists during the COVID-19 disease course, even after the viral clearance. Moreover, previous studies have revealed that some recovered patients were re-detectable positive for SARS-CoV-2 RNA after discharge44. The cause of re-detectable positive remains unclear. One possibility is that a prolonged detrimental effect on the microbiomes exerted by SARS-CoV-2 infection persists even after discharge, which might render convalescent patients susceptible to the residual viremia or reinfection via a more long-lasting dysfunction of the immune system. The microbiome recovery at the taxonomy level seemed to be slowly healed in the oral samples as the PT(−) group shifts towards the healthy controls group based on the dissimilarity distance. In contrast, in the gut samples, the PF(−) group shifts even away from the healthy controls (Fig. 4b). The inconsistency may be due to the period span of sample collection or microbiome characteristics within the oral and gut samples. In a previous metagenomics study, Zuo and colleagues suggested four negatively associated Bacteroides species could be involved in the downregulation of ACE2 expression, and positively regulated E. bacterium 2_2_44A may promote augmenting SARS-CoV-2 infection in the gut17. However, a similar analysis method in our study did not identify the same agents to be associated with the viral load of SARS-CoV-2 (Fig. 4c). Thus, we urge validation of the results that should be taken before recognizing the results.
It is well accepted that unnecessary use of antibiotics should be avoided in the episode of acute respiratory infections as antibiotics have no role in treatment. In reality, 67.9% of patients in our study were treated with antibiotics to prevent potential secondary bacterial co-infections at the early phase of the pandemic. Besides, antibiotics prescribing was elevated in patients with severe conditions as compared to non-severe patients. In a meta-analysis study, Langford has reported that 74.6% of COVID-19 patients received antimicrobial treatment28. Antibiotics treatment can not only eliminate pathogens but commensal microorganisms indiscriminately, which may lead to microbiota dysbiosis and antimicrobial resistance. To circumvent and minimize the effect of antibiotics on the microbiome analysis, we employed a method to count antibiotics treatment as an adjusting variable in the model or to stratify the data for only patients without antibiotics treatment. In addition, unsupervised dimensionality reduction results demonstrated that COVID-19 patients with and without antibiotics treatment shared similar bacterial community structures with a declined diversity in both oral and gut microbiome, shifting away from healthy individuals. It seems that the impacts derived from infection are overwhelming over that of the antibiotic treatment that may apply to the microbiome. The taxa differential analysis also supported this finding, that the differential taxa between the controls and the antibiotics-free patients were largely overlapped with those identified between the controls and all patients (Supplementary Tables 3 and 5).
Collectively, we reported the alterations in both oral and gut microbiomes of SARS-CoV-2 infected patients during hospitalization and made comprehensive analyses to evaluate their potential consequences and implication. The associations between microbial species with disease severity and viral load in patients have suggested the potential of microbiome-based intervention in the prevention and treatment of COVID-19. We believe that the data provides new knowledge with innovative perspectives for tackling and managing the ongoing COVID-19 pandemic.
Methods
Study subject and sample collection
A total of 53 COVID-19 patients and 76 healthy individuals were included in this study. Patients with suspected SARS-CoV-2 infection were confirmed after two sequential positive respiratory tract sample real-time RT-PCR results. Patients were kept hospitalized and under strict observation until the virus was completely eliminated in both respirational and intestinal territories by real-time RT-PCR results. Depending on the sample availability, serial samples were collected from a patient throughout his/her hospitalization period. More specifically, throat swab and fecal samples were collected in both the positive viral RNA test period (P-VRTP, defined as the period of positive nucleic acid tests until the first day of continuous negative tests) and the negative viral RNA test period (N-VRTP, defined as the interval between the first day of negative nucleic acid test until the hospital discharge) for both sample types. Fecal samples were collected from patients who were ever detected with viral RNA in their feces. Only one sample, either throat swab or fecal specimen, was collected from healthy individuals during their physical examination. None of the COVID-19 patients was received antibiotics nor probiotics within 8 weeks before the infection, and none of the healthy individuals was either before this study recruitment. Patients were categorized into two groups based on disease severity: the non-severe group (mild/moderate) and the severe group (severe/critical) following the instruction of the New Coronavirus Pneumonia Prevention and Control Program (7th edition) published by the National Health Commission of China. The demographic information, underlying diseases, clinical indexes, and treatments were summarized from official patients’ medical records. This study was reviewed and approved by the Medical Ethical Committee of the Fifth Affiliated Hospital of Sun Yat-Sen University (approval # K162-1). Written informed consent was obtained from each enrolled subject.
Sample library preparation and sequencing
Samples were inactivated at 56 °C for 30 min before DNA extraction. Extraction of nucleic acids was performed with the CFDA approved nucleic acid extraction kits (QIAamp Viral RNA Mini Kit, Catalog #: 52904, QIAGEN). The concentration and the purity were measured using the NanoDrop One (ThermoFisher Scientific, MA, USA). Sequencing libraries were generated using NEBNext® UltraTM II DNA Library Prep Kit for Illumina (New England Biolabs, MA, USA) following manufacturer’s recommendations, and index codes were added. The universal primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used for amplification of the V3–V4 region of the bacterial 16S rRNA gene. DNA libraries were generated from PCR amplicons targeting the hypervariable regions V3–V4 of the bacterial 16S rRNA gene. After quality assessment with the Qubit 2.0 Fluorometer (ThermoFisher Scientific, MA, USA), the library was then sequenced on an Illumina NovaSeq 6000 platform, and a minimum of 50,000 of 250 bp paired-end reads was generated for the samples.
Bioinformatics analysis
Raw FASTQ files were demultiplexed using the QIIME 2 demux plugin based on their unique barcodes45. Demultiplexed sequences from each sample were stitched, quality filtered, trimmed, de-noised, and then the chimeric sequences were identified and removed using the QIIME 2 dada2 plugin to obtain the feature table46. The QIIME 2 feature-classifier plugin was then used to align feature sequences to a pre-trained GREENGENES 13_8 99% database to generate the taxonomy table47. The data was rarefied prior to alpha and beta diversity analysis using a depth of 13,111 reads. Any contaminating mitochondrial and chloroplast sequences were filtered using the QIIME 2 feature-table plugin. Diversity metrics were calculated and plotted using the core-diversity plugin and the emperor plugin within QIIME 248. The beta diversity significance among groups was examined with PERMANOVA and PERMDISP tests using QIIME 2 plugins and Vegan package in R (version 4.0.2). The differences in the relative abundance of taxa between the patients and healthy control groups were identified in both sample types separately using the linear discriminant analysis effect size (LEfSe)49. Given the possible confounding impact, the output from LEfSe was further validated with Multivariate Association with Linear Models (MaAsLin2) by adjusting confounding factors (age, sex, antibiotic usage, PCR detection result, and patient ID), so only the bacteria taxa agreed by both methods were presented50. PICRUSt2 was used to predict the microbial metabolic pathways to assess the potential functional implication51. QIIME 2 Procrustes plugin was used to examine the fitness of the functional properties and the bacterial composition in the microbiome community with the Procrustes plot, and the correlation between functional properties (Bray Curtis distance) and the bacterial composition (unweighted UniFrac distance) in the microbiome community was examined by two-sided Mantel test.
Detection of SARS-CoV-2 viral load
SARS-CoV-2 viral loads in the throat and fecal swabs were measured using a real-time RT-PCR assay. Viral RNA from throat swabs and fecal samples were extracted using QIAamp Viral RNA Mini Kit (QIAamp Viral RNA Mini Kit, Catalog #: 52904, QIAGEN). Up to 0.1 g of stool or throat swab was suspended in a 2 mL viral transport medium (in 1:10 dilution), followed by centrifugation at 3000 × g for 30 min. The aliquot of the filtrate was used as the starting material. The real-time RT-PCR was carried out with the Novel Coronavirus (2019-nCoV) real-time RT-PCR kit from LifeRiver Ltd. (Catalog #: RR-0479-02). Nucleocapsid gene (N), membrane gene (E), and RNA dependent RNA polymerase gene (RdRp) were the three targeted genes simultaneously amplified and tested. According to the manufacturer’s instructions, a combined result of the three SARS-CoV-2 viral gene targets was used to yield a positive result. Samples were considered negative if the cycle threshold values exceeded 43 cycles. Plasmids containing the full N gene and E gene were obtained to assess SARS-CoV-2 viral copy in the samples (PCDNA6B-SARS-CoV-2-N and PCDNA6B-SARS-CoV-2-E, gifts from Peihui Wang, Cheeloo College of Medicine, Shandong University). Serial 10-fold dilutions of known copies of these plasmids were prepared separately for generating the standard curve. The Ct values of real-time RT-PCR from patient samples were converted into viral RNA copies based on a standard curve.
Statistical analysis
Categorical variables were presented as numbers and percentages (n/N, %) whereas continuous variables were reported as median and interquartile ranges (IQR). According to the distribution of data sets, the one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test or the Kruskal–Wallis test followed by Dunn’s multiple comparisons test were performed using Prism 8.3.0 (GraphPad Software). Given the nature of compositional data for the 16S microbial relative abundance information, we implemented CCLasso to define the correlation between the longitudinal viral load and the timepoint-matched relative abundances of different taxa in the throat swab and fecal samples separately52. The Spearman rank test was used to calculate the correlation between the viral load represented by the abundances of the N gene and the E gene, as their values did not follow the normal distribution according to the Kolmogorov–Smirnov test. All statistical tests were two-sided, and a p-value, of <0.05 was considered significant. Statistical analysis was performed using SPSS version 25.0 (SPSS Inc). Additional custom R scripts were used to making the plots.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Raw FASTQ files of 16S rRNA gene sequences were archived in the Sequence Read Archive (SRA) under Bioproject accession number PRJNA684070. All supporting data are publicly available at https://github.com/XiaominCheng/COVID19-16S.
Code availability
The code used for analyses is publicly available at https://github.com/XiaominCheng/COVID19-16S.
Change history
15 December 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41522-021-00262-z
References
Gheblawi, M. et al. Angiotensin-converting enzyme 2: SARS-CoV-2 Receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 126, 1456–1474 (2020).
Saponaro, F. et al. ACE2 in the era of SARS-CoV-2: controversies and novel perspectives. Front. Mol. Biosci. 7, 588618 (2020).
Mueller, A. L., McNamara, M. S. & Sinclair, D. A. Why does COVID-19 disproportionately affect older people? Aging 12, 9959–9981 (2020).
Ni, W. et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 24, 422 (2020).
Wu, Y. et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 5, 434–435 (2020).
Kumari, P. et al. Potential diagnostics and therapeutic approaches in COVID-19. Clin. Chim. Acta 510, 488–497 (2020).
Hodgson, S. H. et al. What defines an efficacious COVID-19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS-CoV-2. Lancet Infect. Dis. 21, e26–e35 (2021).
Bao, L. et al. Oral Microbiome and SARS-CoV-2: Beware of Lung Co-infection. Front. Microbiol. 11, 1840 (2020).
Mammen, M. J., Scannapieco, F. A. & Sethi, S. Oral-lung microbiome interactions in lung diseases. Periodontology 83, 234–241 (2020).
Li, Y. et al. Salivary mycobiome dysbiosis and its potential impact on bacteriome shifts and host immunity in oral lichen planus. Int J. Oral. Sci. 11, 13 (2019).
Budden, K. F. et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15, 55–63 (2017).
Zhang, D. et al. The cross-talk between gut microbiota and lungs in common lung diseases. Front. Microbiol. 11, 301 (2020).
Enaud, R. et al. The Gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front. Cell Infect. Microbiol. 10, 9 (2020).
Xiang, Z. et al. Potential implications of SARS-CoV-2 oral infection in the host microbiota. J. Oral. Microbiol. 13, 1853451 (2020).
Jia, L. et al. Mechanisms of severe mortality-associated bacterial co-infections following influenza virus infection. Front. Cell Infect. Microbiol. 7, 338 (2017).
Kaul, D. et al. Microbiome disturbance and resilience dynamics of the upper respiratory tract during influenza A virus infection. Nat. Commun. 11, 2537 (2020).
Zuo, T. et al. Alterations in gut microbiota of patients with Covid-19 during time of hospitalization. Gastroenterology https://doi.org/10.1053/j.gastro.2020.05.048 (2020).
Lugli, G. A. et al. A microbiome reality check: limitations of in silico-based metagenomic approaches to study complex bacterial communities. Environ. Microbiol. Rep. 11, 840–847 (2019).
Kitamoto, S. et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell 182, 447–462 (2020).
Schmidt, T. S. et al. Extensive transmission of microbes along the gastrointestinal tract. Elife 8, e42693 (2019).
Human Microbiome Project, Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Lucas-Hourani, M. et al. Inhibition of pyrimidine biosynthesis pathway suppresses viral growth through innate immunity. PLoS Pathog. 9, e1003678 (2013).
Jeong, H. W. et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 26, 1520–1524 (2020).
Rooks, M. G. & Garrett, W. S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352 (2016).
Kriss, M., Hazleton, K. Z., Nusbacher, N. M., Martin, C. G. & Lozupone, C. A. Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery. Curr. Opin. Microbiol. 44, 34–40 (2018).
Fischbach, M. A. Microbiome: focus on causation and mechanism. Cell 174, 785–790 (2018).
Lansbury, L., Lim, B., Baskaran, V. & Lim, W. S. Co-infections in people with COVID-19: a systematic review and meta-analysis. SSRN Electron. J. https://doi.org/10.2139/ssrn.3594598 (2020).
Langford, B. J. et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin. Microbiol. Infect. https://doi.org/10.1016/j.cmi.2020.12.018 (2021).
Sahni, V. & Gupta, S. COVID-19 & periodontitis: the cytokine connection. Med. Hypotheses 144, 109908 (2020).
Yagupsky, P. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect. Dis. 4, 358–367 (2004).
Kosikowska, U., Rybojad, P., Stepien-Pysniak, D., Zbikowska, A. & Malm, A. Changes in the prevalence and biofilm formation of Haemophilus influenzae and Haemophilus parainfluenzae from the respiratory microbiota of patients with sarcoidosis. BMC Infect. Dis. 16, 449 (2016).
Weyand, N. J. Neisseria models of infection and persistence in the upper respiratory tract. Pathog. Dis. 75, 1–13 (2017).
Donati, C. et al. Uncovering oral Neisseria tropism and persistence using metagenomic sequencing. Nat. Microbiol. 1, 16070 (2016).
Gonzalez Navarro, B., Jane Salas, E., Estrugo Devesa, A., Lopez Lopez, J. & Vinas, M. Bacteremia associated with oral surgery: a review. J. Evid. Based Dent. Pr. 17, 190–204 (2017).
Sampson, V., Kamona, N. & Sampson, A. Could there be a link between oral hygiene and the severity of SARS-CoV-2 infections? Br. Dent. J. 228, 971–975 (2020).
Patel, J. & Sampson, V. The role of oral bacteria in COVID-19. Lancet Microbe 1, e105 (2020).
Gu, S. et al. Alterations of the gut microbiota in patients with COVID-19 or H1N1 influenza. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa709 (2020).
Prodan, A., Levin, E. & Nieuwdorp, M. Does disease start in the mouth, the gut or both? Elife 8, e45931 (2019).
Wu, L. et al. SARS-CoV-2 and cardiovascular complications: from molecular mechanisms to pharmaceutical management. Biochem. Pharm. 178, 114114 (2020).
Marotz, C. et al. SARS-CoV-2 detection status associates with bacterial community composition in patients and the hospital environment. Microbiome 9, 1–15 (2021).
Machado, A. S. et al. Oral probiotic bifidobacterium longum supplementation improves metabolic parameters and alters the expression of the renin-angiotensin system in obese mice liver. Biol. Res. Nurs. 23, 100–108 (2021).
Martino, C. et al. Bacterial modification of the host glycosaminoglycan heparan sulfate modulates SARS-CoV-2 infectivity. Preprint at https://biorxiv.org/content/10.1101/2020.08.17.238444v1 (2020).
Eng, A. & Borenstein, E. Taxa-function robustness in microbial communities. Microbiome 6, 45 (2018).
Tao, W. et al. Re-detectable positive SARS-CoV-2 RNA tests in patients who recovered from COVID-19 with intestinal infection. Protein Cell https://doi.org/10.1007/s13238-020-00778-8 (2020).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90 (2018).
Vazquez-Baeza, Y., Pirrung, M., Gonzalez, A. & Knight, R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2, 16 (2013).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. Preprint at https://biorxiv.org/content/10.1101/2021.01.20.427420v1 (2021).
Douglas, G. M. et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688 (2020).
Fang, H., Huang, C., Zhao, H. & Deng, M. CCLasso: correlation inference for compositional data through Lasso. Bioinformatics 31, 3172–3180 (2015).
Acknowledgements
We thank Peihui Wang from Cheeloo College of Medicine, Shandong University for providing PCDNA6B-SARS-CoV-2-N and PCDNA6B-SARS-CoV-2-E. We also thank Dr. Zulfiqar Sahito from Zhejiang University, Dr. Muhammad Younis from the Fifth Affiliated Hospital of Sun Yat-sen University for their help in the language editing. We are thankful to all the study participants. This work was supported by grants from the National Natural Science Foundation of China (82072062), National Science and Technology Key Projects for Major Infectious Diseases (2017ZX10302301-002), Guangzhou Science and Technology Planning Project (201704020226 and 201604020006), National Key Research and Development Program of China (2016YFC1200105), The Three Major Scientific Research Projects of Sun Yat-sen University (20200326236), Guangdong Scientific and Technological Research Project for COVID-19 containment (2020A111128022, 2020B111112003), Guangdong Scientific and Technological Research for COVID-19 (202020012612200001), Zhuhai Scientific and Technological Research Project for COVID-19 containment (ZH22036302200029PWC).
Author information
Authors and Affiliations
Contributions
Y.W., X.C. and G.J. contributed equally to this work. Conceptualization: X.H., H.S., C.G. and J.L.; Funding acquisition: X.H. and H.S.; investigation and lab work: Y.W., G.J., H.T., S.M. and L.T.; microbiota data collection: G.J., H.T. and Y.W.; project administration: Y.W. and X.H.; supervision: X.H., H.S., C.G. and J.L.; bioinformatics and biostatistical analysis: C.G. and X.M.C; writing—original draft: Y.W., X.C. and G.M.J.; writing—review and editing: all. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, Y., Cheng, X., Jiang, G. et al. Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization. npj Biofilms Microbiomes 7, 61 (2021). https://doi.org/10.1038/s41522-021-00232-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-021-00232-5
This article is cited by
-
Standardization of the protocol for oral cavity examination and collecting of the biological samples for microbiome research using the next-generation sequencing (NGS): own experience with the COVID-19 patients
Scientific Reports (2024)
-
Assessment of microbiota in the gut and upper respiratory tract associated with SARS-CoV-2 infection
Microbiome (2023)
-
Gut microbiota composition during hospitalization is associated with 60-day mortality after severe COVID-19
Critical Care (2023)
-
Gut microbiota in COVID-19: key microbial changes, potential mechanisms and clinical applications
Nature Reviews Gastroenterology & Hepatology (2023)
-
The Upper Respiratory Tract Microbiome Network Impacted by SARS-CoV-2
Microbial Ecology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.