Abstract
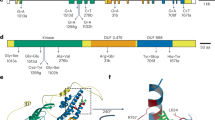
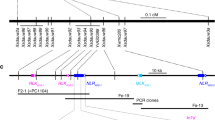
Stem rust caused by the fungus Puccinia graminis f. sp. tritici (Pgt) is a devastating disease of the global staple crop wheat. Although this disease was largely controlled in the latter half of the twentieth century, new virulent strains of Pgt, such as Ug99, have recently evolved1,2. These strains have caused notable losses worldwide and their continued spread threatens global wheat production. Breeding for disease resistance provides the most cost-effective control of wheat rust diseases3. A number of rust resistance genes have been characterized in wheat and most encode immune receptors of the nucleotide-binding leucine-rich repeat (NLR) class4, which recognize pathogen effector proteins known as avirulence (Avr) proteins5. However, only two Avr genes have been identified in Pgt so far, AvrSr35 and AvrSr50 (refs. 6,7), and none in other cereal rusts8,9. The Sr27 resistance gene was first identified in a wheat line carrying an introgression of the 3R chromosome from Imperial rye10. Although not deployed widely in wheat, Sr27 is widespread in the artificial crop species Triticosecale (triticale), which is a wheat–rye hybrid and is a host for Pgt11,12. Sr27 is effective against Ug99 (ref. 13) and other recent Pgt strains14,15. Here, we identify both the Sr27 gene in wheat and the corresponding AvrSr27 gene in Pgt and show that virulence to Sr27 can arise experimentally and in the field through deletion mutations, copy number variation and expression level polymorphisms at the AvrSr27 locus.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All sequence data from this study are available in NCBI under BioProject PRJNA695305 (Sr27 MutRenSeq data) and PRJNA698655 (Pgt21-0 mutants). This includes raw data for Fig. 2b and Supplementary Figs. 2 and 8. Figure 3 is derived from raw sequence data available in NCBI BioProject PRJNA253722 and PRJNA415866. All other relevant data are available on request from the corresponding author.
Code availability
Scripts and files for MutRenSeq analysis are available at https://github.com/TC-Hewitt/MuTrigo.
References
Singh, R. P. et al. Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105, 872–884 (2015).
Li, F. et al. Emergence of the Ug99 lineage of the wheat stem rust pathogen through somatic hybridisation. Nat. Commun. 10, 5068 (2019).
Ellis, J. G., Lagudah, E. S., Spielmeyer, W. & Dodds, P. N. The past, present and future of breeding rust resistant wheat. Front. Plant Sci. https://doi.org/10.3389/fpls.2014.00641 (2014).
Periyannan, S., Milne, R. J., Figueroa, M., Lagudah, E. S. & Dodds, P. N. An overview of genetic rust resistance: from broad to specific mechanisms. PloS Pathog. https://doi.org/10.1371/journal.ppat.1006380 (2017).
Garnica, D. P., Nemri, A., Upadhyaya, N. M., Rathjen, J. P. & Dodds, P. N. The ins and outs of rust haustoria. PloS Pathog. https://doi.org/10.1371/journal.ppat.1004329 (2014).
Chen, J. et al. Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 358, 1607–1610 (2017).
Salcedo, A. et al. Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 358, 1604–1606 (2017).
Bakkeren, G. & Szabo, L. J. Progress on molecular genetics and manipulation of rust fungi. Phytopathology 110, 532–543 (2020).
Figueroa, M., Dodds, P. N. & Henningsen, E. C. Evolution of virulence in rust fungi—multiple solutions to one problem. Curr. Opin. Plant Biol. 56, 20–27 (2020).
McIntosh, R. A., Wellings, C. R. & Park, R. F. Wheat Rusts: An Atlas of Resistance Genes (CSIRO Publications, 1995).
McIntosh, R. A., Luig, N. H., Milne, D. L. & Cusick, J. Vulnerability of triticales to wheat stem rust. Can. J. Plant. Pathol. 5, 61–69 (1983).
Zhang, J., Wellings, C. R., McIntosh, R. A. & Park, R. F. Seedling resistances to rust diseases in international triticale germplasm. Crop Pasture Sci. 61, 1036–1048 (2010).
Jin, Y., Pretorius, Z. A. & Singh, R. P. New virulence within race TTKS (Ug99) of the stem rust pathogen and effective resistance genes. Phytopathology 97, S137–S137 (2007).
Olivera, P. et al. Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013–14. Phytopathology 105, 917–928 (2015).
Olivera, P. D. et al. Presence of a sexual population of Puccinia graminis f. sp. tritici in Georgia provides a hotspot for genotypic and phenotypic diversity. Phytopathology 109, 2152–2160 (2019).
Upadhyaya, N. M. et al. Comparative genomics of Australian isolates of the wheat stem rust pathogen Puccinia graminis f. sp. tritici reveals extensive polymorphism in candidate effector genes. Front. Plant Sci. https://doi.org/10.3389/fpls.2014.00759 (2015).
Zhang, J., Zhang, P., Karaoglu, H. & Park, R. F. Molecular characterization of australian isolates of Puccinia graminis f. sp. tritici supports long-term clonality but also reveals cryptic genetic variation. Phytopathology 107, 1032–1038 (2017).
Lewis, C. M. et al. Potential for re-emergence of wheat stem rust in the United Kingdom. Commun. Biol. 1, 13 (2018).
Visser, B. et al. Microsatellite analysis and urediniospore dispersal simulations support the movement of Puccinia graminis f. sp. tritici from Southern Africa to Australia. Phytopathology 109, 133–144 (2019).
Visser, B., Herselman, L. & Pretorius, Z. A. Genetic comparison of Ug99 with selected South African races of Puccinia graminis f. sp. tritici. Mol. Plant Pathol. 10, 213–222 (2009).
Lee, W. S., Hammond-Kosack, K. E. & Kanyuka, K. Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 160, 582–590 (2012).
Qutob, D. et al. Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS ONE 4, e5066 (2009).
Pais, M. et al. Gene expression polymorphism underpins evasion of host immunity in an asexual lineage of the Irish potato famine pathogen. BMC Evol. Biol. 18, 93 (2018).
Wu, W. et al. Flax rust infection transcriptomics reveals a transcriptional profile that may be indicative for rust Avr genes. PLoS ONE 4, e0226106 (2019).
Steuernagel, B. et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655 (2016).
Appels, R. et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191 (2018).
Saur, I. M. L., Bauer, S., Lu, X. L. & Schulze-Lefert, P. A cell death assay in barley and wheat protoplasts for identification and validation of matching pathogen AVR effector and plant NLR immune receptors. Plant Methods https://doi.org/10.1186/s13007-019-0502-0 (2019).
Luo, M. et al. A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. 39, 561–566 (2020).
Mago, R. et al. The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat. Plants 1, 15186 (2015).
Marais, G. F. An evaluation of three Sr27-carrying wheat × rye translocations. South Afr. J. Plant Soil 18, 135–136 (2001).
Mago, R. et al. Transfer of stem rust resistance gene SrB from Thinopyrum ponticum into wheat and development of a closely linked PCR-based marker. Theor. Appl Genet. 132, 371–382 (2019).
Zhang, J. et al. A recombined Sr26 and Sr61 disease resistance gene stack in wheat encodes unrelated NLR genes. Nat. Commun. 12, 3378 (2021).
Wulff, B. B., Horvath, D. M. & Ward, E. R. Improving immunity in crops: new tactics in an old game. Curr. Opin. Plant Biol. 14, 468–476 (2011).
Hubbard, A. et al. Field pathogenomics reveals the emergence of a diverse wheat yellow rust population. Genome Biol. 16, 23 (2015).
Sperschneider, J. et al. The stem rust fungus Puccinia graminis f. sp. tritici induces centromeric small RNAs during late infection that direct genome-wide DNA methylation. Preprint at bioRxiv https://doi.org/10.1101/469338 (2020).
Park, R. F. Stem rust of wheat in Australia. Aust. J. Agric. Res. 58, 558–566 (2007).
Singh, S. J. & Mcintosh, R. A. Allelism of 2 genes for stem rust resistance in triticale. Euphytica 38, 185–189 (1988).
Acosta, A. C. The transfer of stem rust resistance from rye to wheat. Dissertation Abstr. 23, 34–35 (1962).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint at https://arxiv.org/abs/1207.3907 (2012).
Stamatakis, A., Ludwig, T. & Meier, H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21, 456–463 (2005).
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Heibl, C. PHYLOCH: R language tree plotting tools and interfaces to diverse phylogenetic software packages (R Foundation for Statistical Computing, 2008); http://www.christophheibl.de/Rpackages.html
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Yin, T., Cook, D. & Lawrence, M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 13, R77 (2012).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evolution 35, 1547–1549 (2018).
Lee, W. S., Rudd, J. J. & Kanyuka, K. Virus induced gene silencing (VIGS) for functional analysis of wheat genes involved in Zymoseptoria tritici susceptibility and resistance. Fungal Genet. Biol. 79, 84–88 (2015).
Franco-Orozco, B. et al. A new proteinaceous pathogen-associated molecular pattern (PAMP) identified in Ascomycete fungi induces cell death in Solanaceae. New Phytol. 214, 1657–1672 (2017).
Hen-Avivi, S. et al. A metabolic gene cluster in the wheat W1 and the barley cer-cqu loci determines beta-diketone biosynthesis and glaucousness. Plant Cell 28, 1440–1460 (2016).
Lee, W. S., Rudd, J. J., Hammÿond-Kosack, K. E. & Kanyuka, K. Mycosphaerella graminicola LysM effector-mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol. Plant Microbe Interact. 27, 236–243 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Mago, R. et al. Generation of loss-of-function mutants for wheat rust disease resistance gene cloning. Wheat Rust. Dis.: Methods Protoc. 1659, 199–205 (2017).
Mago, R. et al. Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor. Appl. Genet. 111, 496–504 (2005).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Rabanus-Wallace, M. T. et al. Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet. 53, 564–573 (2021).
Akamatsu, A. et al. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13, 465–476 (2013).
Arndell, T. et al. gRNA validation for wheat genome editing with the CRISPR-Cas9 system. BMC Biotech. 19, 71 (2019).
Bemoux, M. et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9, 200–211 (2011).
Cesari, S. et al. Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC-NLR proteins. Proc. Natl Acad. Sci. USA 113, 10204–10209 (2016).
Cesari, S. et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481 (2013).
Richardson, T., Thistleton, J., Higgins, T. J., Howitt, C. & Ayliffe, M. Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell, Tissue Organ Cult. 119, 647–659 (2014).
Acknowledgements
Work described here was supported by funding from the 2Blades foundation to P.N.D. J.C. was supported by a Chinese Scholarship Council postgraduate fellowship. J.S. was supported by an Australian Research Council Discovery Early Career Researcher Award (no. DE190100066). K.K. and V.P. acknowledge financial support by the Institute Strategic Program Grant ‘Designing Future Wheat’ (grant no. BB/P016855/1) from the Biotechnology and Biological Sciences Research Council of the UK. H.N.-P. was supported by USDA-NIFA grant no. 2018-67013-27819.
Author information
Authors and Affiliations
Contributions
P.N.D. conceptualized the project, acquired funding and supervised the work. N.M.U., R.M., V.P., M.L., A.W., D.O., J.C., J.Z., D.B., M.A. and L.H. acquired experimental data. N.M.U., R.M., J.S., H.N.-P., T.H., M.F., K.K., J.G.E. and P.N.D conducted data analysis. P.N.D. drafted the manuscript and all authors contributed to review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Beat Keller, Simon Krattinger and Brande Wulff for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Tables 1 and 2 and source data as uncropped gel/blot images for Supplementary Figs. 3, 5a, 6 and 11.
Rights and permissions
About this article
Cite this article
Upadhyaya, N.M., Mago, R., Panwar, V. et al. Genomics accelerated isolation of a new stem rust avirulence gene–wheat resistance gene pair. Nat. Plants 7, 1220–1228 (2021). https://doi.org/10.1038/s41477-021-00971-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00971-5
This article is cited by
-
Pooled effector library screening in protoplasts rapidly identifies novel Avr genes
Nature Plants (2024)
-
Exploratory genomic sequence analysis reveals structural differences at key loci for growth habit, seed dormancy, and rust resistance in barley
Genetic Resources and Crop Evolution (2024)
-
The broad use of the Pm8 resistance gene in wheat resulted in hypermutation of the AvrPm8 gene in the powdery mildew pathogen
BMC Biology (2023)
-
Conservatively transmitted alleles of key agronomic genes provide insights into the genetic basis of founder parents in bread wheat (Triticum aestivum L.)
BMC Plant Biology (2023)
-
Fighting wheat rusts in China: a look back and into the future
Phytopathology Research (2023)