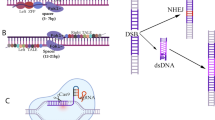
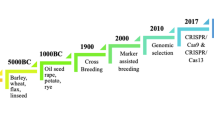
Abstract
The recent development of gene editing has generated a lot of excitement for the future of crop improvement. At a technical level, targeted gene modification through gene editing (GE) provides an opportunity to build on natural variation to target genetic changes for the creation of new varieties, crops, and market opportunities. Specialty crops, represented by hundreds of plant species, are poised to take advantage of GE and provide solutions to impact traits of importance to the consumer (such as flavor and nutrition), the environment (such as reducing waste, and transport distances), and the farmer (yield and pest/disease resistance). However, many specialty crops face challenges to implement GE such as complex uncharacterized genomes, polyploidy, and long breeding and production cycles. A small number of specialty crops such as tomato are already taking advantage of GE and can represent a blueprint for continued improvement of important specialty crops some of which have gone for centuries without significant genetic improvement.

Similar content being viewed by others
References
Antcliff A, Webster W (1962) Bruce’s sport — a mutant of the sultana. Anim Prod Sci 2:97–100
Armstrong and Lane (2019) Genetically modified reduced browning, fruit producing plant and produced fruit thereof, and method of obtaining such. US Patent 10,227,602 B2
Bai Y, Lindhout P (2007) Domestication and breeding of tomatoes: what have we gained and what can we gain in the future? Ann Bot 100:1085–1094
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell Online 26:151–163
Baltes NJ, Gil-Humanes J, Voytas DF (2017) Chapter one genome engineering and agriculture: opportunities and challenges. Prog Mol Biol Transl 149:1–26
Baranski R, Klimek-Chodacka M, Lukasiewicz A (2019) Approved genetically modified (GM) horticultural plants: a 25-year perspective. Folia Hortic 31:3–49
Beacham AM, Vickers LH, Monaghan JM (2019) Vertical farming: a summary of approaches to growing skywards. J Hortic Sci Biotechnol 94:1–7
Bharat SS, Li S, Li J, Yan L, Xia L (2019) Base editing in plants: current status and challenges. Crop J 8:384–395. https://doi.org/10.1016/j.cj.2019.10.002
Brooks C, Nekrasov V, Lippman ZB, Eck JV (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-Associated9 system. Plant Physiol 166:1292–1297
Čermák T, Baltes, NJ, Čegan R, Zhang Y, Voytas DF (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16:1–15
Charrier A, Vergne E, Dousset N, Richer A, Petiteau A, Chevreau E (2019) Efficient targeted mutagenesis in apple and first time editing of pear ssing the CRISPR-Cas9 System. Front Plant Sci 10:40
Chu Y, Jang J, Huang Z, van der Knaap E (2019) Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct 3:e00142
Costa LD, Piazza S, Pompili V, Salvagnin U, Cestaro A, Moffa L, Vittani L, Moser C, Malnoy M (2020) Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci Rep 10:20155
Danilevicz MF, Fernandez CGT, Marsh JI, Bayer PE, Edwards D (2020) Plant pangenomics: approaches, applications and advancements. Curr Opin Plant Biol 54:18–25
de T Thomazella DP, Brail Q, Dahlbeck D, Staskawicz B (2016) CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. BioRxiv, 06482. https://doi.org/10.1101/064824
Demirer GS, Zhang H, Goh NS, González-Grandío E Landry MP (2019) Carbon nanotube–mediated DNA delivery without transgene integration in intact plants. Nature Protocols 14:2954–2971
Doebley JF, Gaut BS, Thomazella BD (2006) The molecular genetics of crop domestication. Cell 127:1309–1321
Enfissi EMA, Nogueira M, Bramley PM, Fraser PD (2017) The regulation of carotenoid formation in tomato fruit. Plant J 89:774–788
Eng WH, Ho WS (2019) Polyploidization using colchicine in horticultural plants: A review. Sci Hortic-amsterdam 246: 604–617
FAO (2019) The state of the world’s biodiversity for food and agriculture, J. Bélanger & D. Pilling (eds.). FAO Commission on Genetic Resources for Food and Agriculture Assessments. Rome. 572 pp. (http://www.fao.org/3/CA3129EN/CA3129EN.pdf)
Fernie AR, Yan J (2020) SPOTLIGHT: Targeting key genes to tailor old and new crops for a greener agriculture. Mol Plant 13:354–356
Flachowsky H, Roux PL, Peil A, Patocchi A, Richter K, Hanke M (2011) Application of a high-speed breeding technology to apple (Malus × domestica) based on transgenic early flowering plants and marker-assisted selection. New Phytol 192:364–377
Fräbel S, Lawit SJ, Nie J, Schwark DG, Poorten TJ, Graham ND (2021) Progress in precise and predictable genome editing in plants with base editing. In Genome editing for precision crop breeding (pp. 123–146). Burleigh Dodds Science Publishing
Gallego-Bartolomé J, Gardiner J, Liu W, Papikian A, Ghoshal B, Kuo HY, Zhao JM-C, Segal DJ, Jacobsen SE (2018) Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc Natl Acad Sci 115:2125–2134
Galli M, Gallavotti A (2016) Expanding the regulatory network for meristem size in plants. Trends Genet 32:372–383
Gaston A, Osorio S, Denoyes B, Rothan C (2020) Applying the Solanaceae strategies to strawberry crop improvement. Trends Plant Sci 25:130–140
Hake S, Ross-Ibarra J (2015) Genetic, evolutionary and plant breeding insights from the domestication of maize. Elife 4:e05861
Hao M, Zhang L, Ning S, Huang L, Yuan Z, Wu B, Yan Z, Dai S, Jiang B, Zheng Y, Liu D (2020) The resurgence of introgression breeding, as exemplified in wheat improvement. Front Plant Sci 11:252
Hare PD, Chua N-H (2002) Excision of selectable marker genes from transgenic plants. Nat Biotechnol 20:575–580
Heuvelink E (2005) Developmental processes. Crop Production Science in Horticulture 13: 53–83
Hollender CA, Geretz AC, Slovin JP, Liu Z (2012) Flower and early fruit development in a diploid strawberry, Fragaria vesca. Planta 235:1123–1139
Huang T-K, Puchta H (2019) CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep 38:443–453
Huang TK, Puchta H (2021) Novel CRISPR/Cas applications in plants: from prime editing to chromosome engineering. Transgenic Res 1–21 https://doi.org/10.1007/s11248-021-00238-x
Khan MZ, Zaidi SS, Amin I, Mansoor S (2019) A CRISPR way for fast-forward crop domestication. Trends Plant Sci:24293–24296
Kharkwal MC, Pandey RN, Pawar SE (2004) Mutation breeding for crop improvement. In: Jain HK, Kharkwal MC (eds) Plant Breeding. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-1040-5_26
Klap C, Yeshayahou E, Bolger AM, Arazi T, Gupta SK, Shabtai S, Usadel B, Salts Y, Barg R (2017) Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol J 15:634–647
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424
Kwon C-T, Heo J, Lemmon ZH, Capua Y, Hutton SF, Eck JV, Park SJ, Lippman ZB (2019) Rapid customization of Solanaceae fruit crops for urban agriculture. Nat Biotechnol 38:182–188
Kyriakidou M, Tai HH, Anglin NL, Ellis D Strömvik MV (2018) Current strategies of polyploid plant genome sequence assembly. Front Plant Sci 9:1660
Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, Rodriguez-Leal D, Eck JV, Lippman ZB (2018) Rapid improvement of domestication traits in an orphan crop by genome editing. Nat Plant 4:766–770
Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C (2018) Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol 36:1160–1163
Lindbo JA, Dougherty WG (2005) Plant pathology and RNAi: a brief history. Ann Rev Phytopathol 43:191–204
Lor VS, Starker CG, Voytas DF, Weiss D, Olszewski NE (2014) Targeted mutagenesis of the tomato PROCERA gene using transcription activator-like effector nucleases. Plant Physiol 166:1288–1291
Lu Y, Tian Y, Shen R, Yao Q, Zhong D, Zhang X, Zhu J (2020) Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol J 19:415–417
Lu Y, Zhu JK (2017) Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol Plant 10:523–525
Malabarba J, Chevreau E, Dousset N, Veillet F, Moizan J, Vergne E (2020) New strategies to overcome present CRISPR/Cas9 limitations in apple and pear: efficient dechimerization and base editing. Int J Mol Sci 22:319
Malnoy M, Viola R, Jung M-H, Koo O-J, Kim S, Kim J-S, Velasco R, Kanchiswamy CN (2016) DNA-Free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci 7:1904
Martínez MIS, Bracuto V, Koseoglou E, Appiano M, Jacobsen E, Visser RG, Wolters AMA, Bai Y (2020) CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol 20:1–13
Massel K, Lam Y, Wong ACS, Hickey LT, Borrell AK, Godwin ID (2021) Hotter, drier, CRISPR: the latest edit on climate change. Theor Appl Genet 134:1691–1709. https://doi.org/10.1007/s00122-020-03764-0
Maxmen A (2019) CRISPR might be the banana’s only hope against a deadly fungus. Nature 574:15–15
Meinke DW (2013) A survey of dominant mutations in Arabidopsis thaliana. Trends Plant Sci 18:84–91
Mes PJ, Boches P, Myers JR, Durst R (2008) Characterization of tomatoes expressing anthocyanin in the fruit. J Am Soc Hortic Sci 133:262–269
Mohan C (2016) Genome editing in sugarcane: challenges ahead. Front Plant Sci 7:1542
Muños S, Ranc N, Botton E, Bérard A, Rolland S, Duffé P, Carretero Y, Le Paslier MC, Delalande C, Bouzayen M, Brunel D (2011) Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol 156:2244–2254
Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S (2017) Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep 7:482
Nishitani C, Hirai N, Komori S, Wada M, Okada K, Osakabe K, Yamamoto T, Osakabe Y (2016) Efficient genome editing in apple using a CRISPR/Cas9 system. Sci Rep 6:31481
Nonaka S, Arai C, Takayama M, Matsukura C, Ezura H (2017) Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci Rep 7:7057
Ortigosa A, Gimenez-Ibanez S, Leonhardt N, Solano R (2019) Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol J 17:665–673
Park J, Choe S (2019) DNA-free genome editing with preassembled CRISPR/Cas9 ribonucleoproteins in plants. Transgenic Res 28:61–64
Pompili V, Costa LD, Piazza S, Pindo M, Malnoy M (2019) Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol J 18:845–858. https://doi.org/10.1111/pbi.13253.
Posé S, Paniagua C, Cifuentes M, Blanco-Portales R, Quesada MA, Mercado JA (2013) Insights into the effects of polygalacturonase FaPG1 gene silencing on pectin matrix disassembly, enhanced tissue integrity, and firmness in ripe strawberry fruits. J Exp Bot 64:3803–3815
Rathjen H, Robinson S (1992) Characterisation of a variegated grapevine mutant showing reduced polyphenol oxidase activity. Funct Plant Biol 19:43–54
Razifard H, Ramos A, Valle ALD, Bodary C, Goetz E, Manser EJ, Li X, Zhang L, Visa S, Tieman D et al (2020) Genomic evidence for complex domestication history of the cultivated tomato in Latin America. Mol Biol Evol 37:1118–1132
Ritchie H, Roser M (2017) Crop yields. https://ourworldindata.org/crop-yields
Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171:470–480
Scorza R, Callahan A, Dardick C, Ravelonandro M, Polak J, Malinowski T, Zagrai I, Cambra M, Kamenova I (2013) Genetic engineering of Plum pox virus resistance: ‘HoneySweet’ plum—from concept to product. Plant Cell Tissue Organ Cult 115:1–12
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35:441–443
Shipman EN, Yu J, Zhou J, Albornoz K, Beckles DM (2021) Can gene editing reduce postharvest waste and loss of fruit, vegetables, and ornamentals? Hortic Res 8:1
Silva CJ, van den Abeele C, Ortega-Salazar I, Papin V, Adaskaveg JA, Wang D, Casteel CL, Seymour GB, Blanco-Ulate B (2021) Host susceptibility factors render ripe tomato fruit vulnerable to fungal disease despite active immune responses. J Exp Bot 72:2696–2709
Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Eck JV, Jiménez-Gómez JM, Lippman ZB (2016) Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet 49:162–168
Srinivasan C, Dardick C, Callahan A, Scorza R (2012) Plum (Prunus domestica) trees transformed with poplar FT1 result in altered architecture, dormancy requirement, and continuous flowering. PLoS One 7:e40715
Stoddard TJ, Clasen BM, Baltes NJ, Demorest ZL, Voytas DF, Zhang F, Luo S (2016) Targeted mutagenesis in plant cells through transformation of sequence-specific nuclease mRNA. PLoS One 11:e0154634
Svitashev S, Schwartz C, Lenderts B, Young JK, Cigan AM (2016) Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat Commun 7:1–7
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327:818–822
Toda E, Koiso N, Takebayashi A, Ichikawa M, Kiba T, Osakabe K, Osakabe Y, Sakakibara H, Kato N, Okamoto T (2019) An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat Plants 5:363–368
Ueta R, Abe C, Watanabe T, Sugano SS, Ishihara R, Ezura H, Osakabe Y, Osakabe K (2017) Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci Rep 7:507
USDA (2014) What is a specialty crop. https://www.ams.usda.gov/services/grants/scbgp/specialty-crop
Veitia RA (2007) Exploring the molecular etiology of dominant-negative mutations. Plant Cell 19:3843–3851
Vernyik V, Karcagi I, Tímár E, Nagy I, Györkei Á, Papp B, Györfy Z, Pósfai G (2020) Exploring the fitness benefits of genome reduction in Escherichia coli by a selection-driven approach. Sci Rep 10:7345
Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, Klasić M, Zoldoš V (2016) Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nuc Acid Res 44:5615–5628
Wang T, Zhang H, Zhu H (2019) CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic Res 6:77
Wang Z, Meng D, Wang A, Li T, Jiang S, Cong P, Li T (2013) The methylation of the PcMYB10 promoter Is associated with green-skinned sport in Max Red Bartlett Pear. Plant Physiol 162:885–896
Xu C, Liberatore KL, MacAlister CA, Huang Z, Chu Y-H, Jiang K, Brooks C, Ogawa-Ohnishi M, Xiong G, Pauly M, Van Eck J, Matsubayashi Y, van der Knaap E, Lippman ZB (2015) A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat Genet 47:784–792
Xu X, Hulshoff MS, Tan X, Zeisberg M, Zeisberg EM (2020) CRISPR/Cas Derivatives as novel gene modulating tools: possibilities and in vivo applications. Int J Mol Sci 21:3038
Yang CJ, Samayoa LF, Bradbury PJ, Olukolu BA, Xue W, York AM, Tuholski MR, Wang W, Daskalska LL, Neumeyer MA et al (2019) The genetic architecture of teosinte catalyzed and constrained maize domestication. Proc Natl Acad Sci 116:201820997
Yeager AF (1927) Determinate growth in the tomato. J Hered 18:263–265
Young TR, Firoozabady E (2010) Transgenic pineapple plants with modified carotenoid levels and methods of their production. US Patent US 7.663,021 B2
Yuste-Lisbona FJ, Fernández-Lozano A, Pineda B, Bretones S, Ortíz-Atienza A, García-Sogo B, Müller NA, Angosto T, Capel J, Moreno V, Jiménez-Gómez JM, Lozano R (2020) ENO regulates tomato fruit size through the floral meristem development network. Proc Natl Acad Sci 117:8187–8195
Zhang K, Wang X, Cheng F (2019) Plant Polyploidy: origin, evolution, and its influence on crop domestication. Hortic Plant J 5:231–239
Zhebentyayeva T, Shankar V, Scorza R, Callahan A, Ravelonandro M, Castro S, DeJong T, Saski CA, Dardick C (2019) Genetic characterization of worldwide Prunus domestica (plum) germplasm using sequence-based genotyping. Hortic Res 6:12
Zhou J, Wang G, Liu Z (2018) Efficient genome editing of wild strawberry genes, vector development and validation. Plant Biotechnol J 16:1868–1877
Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LE (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol 36:1211–1216
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: Gary Bannon
Rights and permissions
About this article
Cite this article
Bate, N.J., Dardick, C.D., de Maagd, R.A. et al. Opportunities and challenges applying gene editing to specialty crops. In Vitro Cell.Dev.Biol.-Plant 57, 709–719 (2021). https://doi.org/10.1007/s11627-021-10208-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-021-10208-x