Abstract
Bifidobacterium animalis subsp. lactis Bb12 is a widely used probiotic that provides numerous health benefits to its host, many due to its immunomodulatory properties. Although the precise mechanism of modulation is still under investigation, several reports associate the interaction of TLR2 with components of the bacterial cell wall inducing a signaling cascade that culminates with the production of cytokines and co-stimulatory molecules. MicroRNAs (miRNAs) have emerged as important post-transcriptional regulators of immune responses, including those toward probiotics. In this study, we analyzed the miRNA expression profile in swine monocytes exposed to Bb12 by using an anti-TLR2 blocking strategy and Bb12 involvement in the regulation of the TLR2 pathway. As a result, the expression of 40 miRNAs was influenced by the treatments (p < 0.01), and 15 differentially expressed miRNAs with validated miRNA–mRNA interactions with around 26 proteins related to the TLR2 pathway were identified. The miRNAs upregulated in response to Bb12 included miR-15a-5p, miR-16-5p, miR-26a-5p, miR-29b-3p, and miR-30d-5p, and the following showed downregulation: miR-181a-5p, miR-19b-3p, miR-21-5p, miR-23a-5p, and miR-221-3p. The expression of let-7c-5p, let-7f-5p, miR-146b-5p, miR-150-5p, and miR-155-5p was increased by Bb12 only when TLR2 was blocked. The identified miRNA common targets were downstream proteins from bacterial recognition via TLR2, such as MyD88, TRAF6, and MAPK members; transcription factors such as NF-κB and AP-1; and cytokines such as IL-6, IL-10, and TNF-α. TLR2 participation was abrogated by anti-TLR2 antibody and suggests that bacterial recognition is complemented by other receptors since there were still changes in the microtranscriptome.
Similar content being viewed by others
Introduction
Probiotic microorganisms have been widely studied due to their beneficial effects on the health of vertebrates, including animals. Bifidobacterium animalis subsp. lactis Bb12 (Bb12), one of the most studied probiotic organisms, has been shown to improve intestinal function, to enhance protection against infectious diarrhea, and to reduce the secondary effects of antibiotic treatment [1]. Bb12 has also been shown to be immunomodulatory, inducing the production of interleukin (IL)-10 and moderately IL-12 in immune cells [2]. The main mechanism of immune stimulation by Gram-positive bacteria such as Bb12 is mediated by receptor recognition of bacterial cell membrane components, like peptidoglycans (PGNs) and lipoteichoic acid (LTA), for which eukaryotic cells have specific receptors including toll-like receptor 2 (TLR2). Upon binding, TLR2 recruits myeloid differentiation primary response 88 protein (MyD88), initiating a signaling cascade that leads to the activation of at least two transcription factors, namely, nuclear factor-kappa B (NF-κB) and activator protein 1 (AP-1) [3], which in turn induce the production of cytokines.
The use of probiotics has extended to several species, such as the pig, due to its economic importance and as a model for biomedical studies [4,5,6,7]. We have previously reported that Bb12 induces the production of IL-10, and the mechanism involves recruiting of TLR2 [8]. In the last few years, studies have focused on protein regulation by microRNAs (miRNAs), which are small non-coding RNA molecules ranging from 16 to 27 nucleotides that participate in the regulation of several cellular processes including differentiation and proliferation. Changes in miRNA expression may alter the expression of target proteins at transcriptional and post-transcriptional levels by regulating messenger RNA (mRNA) translation or stability in the cytoplasm, mediated by the binding of a seed region, the most common motif for pairing miRNAs to their target mRNA [9]. Current progress in the development of bioinformatics research tools has provided deeper information about miRNA–mRNA interactions, making it possible to elucidate regulatory mechanisms mediated by miRNAs on immune responses. A single miRNA can regulate the production of hundreds of proteins through their mRNAs, and we hypothesize that the diverse immune responses by probiotics could be regulated in this manner. Therefore, we investigated the role of TLR2 on the miRNA profile induced by Bb12 in swine monocytes, looking to provide an overview of differential miRNA expression that may help to identify key miRNAs involved in the regulation of the TLR2 pathway by Bifidobacterium animalis subsp. lactis Bb12.
Materials and Methods
Bacterial Preparation
Bb12 was grown on Man, Rogosa, and Sharpe (MRS) broth (Merck, Darmstadt, Germany) supplemented with 0.05% cysteine (Sigma-Aldrich, St. Louis MO, USA) at 37 °C for 48 h under anaerobic conditions. Bacterial cultures were centrifuged at 3000 × g for 10 min (centrifuge Sorvall ST16, Thermo Fisher Scientific, Waltham, MA, USA). The bacterial pellet was washed with phosphate buffer saline (PBS; 0.137 M NaCl, 0.0027 M KCl, 0.01 M Na2HPO4, 0.0018 M KH2PO4, pH 7.4), and the bacterial concentration was adjusted to 6 × 108 CFU/mL, corresponding to the turbidity McFarland scale 2.0. All reagents were from Sigma-Aldrich unless otherwise stated.
Animals
Conventional female pigs (2–4 months old) were obtained from a local farm, free of porcine reproductive and respiratory virus, influenza virus, and swine enteric coronavirus diseases. The animals were housed at the animal facility of the Centro de Investigación en Alimentación y Desarrollo (CIAD) with ad libitum access to water and food. The animals were euthanized according to the ethical standards of Mexican regulation NOM‐033‐ZOO‐1995. Animal experimentation was approved by the Ethics Committee of CIAD.
Isolation of Porcine Monocytes
Peripheral blood was collected in tubes with 1.5 mg/mL ethylenediaminetetraacetic acid (EDTA) as an anticoagulant. Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation using Ficoll-Paque Plus (GE Healthcare Bio-Sciences, Uppsala, Sweden). PBMCs were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY, USA) supplemented with 5000 IU/mL penicillin, 5 µg/mL streptomycin (Sigma-Aldrich), and 10% fetal bovine serum (FBS, Gibco). Cells (5 × 107) were placed in a T75 flask (Corning Inc., Corning, NY, USA) and incubated at 37 °C and 5% CO2 for 90 min to allow monocytes to adhere. Flasks were washed four times with PBS to eliminate non-adherent cells. The remaining adherent cells were incubated for 10 min with a trypsin–EDTA solution (Sigma-Aldrich) to detach cells, were further harvested, and washed with DMEM. Cell viability testing was performed with the trypan blue exclusion method, with an average percentage of 95% living cells. Monocyte population was identified with surface cell markers CD172a and CD163 (VMRD, Pullman, WA, USA) by flow cytometry (FACSCanto II, Becton Dickinson Biosciences, San Jose, CA, USA) as previously reported [8].
Cell Stimulation
Monocytes (2 × 106) were added to 48-well plates and incubated either alone (unstimulated control) or with the addition of Bb12 (ratio 1:100 cell/bacteria) for 4 h, which allowed to detect an increase of IL-10 transcript expression. Incubation time was selected according to previously reported results [8]. Groups used in this study were as follows: unstimulated swine monocytes, swine monocytes stimulated with Bb12 for 4 h (group designated “Bb12”), and swine monocytes incubated with anti-TLR2 antibody and Bb12 for 4 h (group designated “anti-TLR2 + Bb12”). For TLR2 blocking experiments, 40 μg/mL of purified anti-mouse/human CD282 (TLR2) antibody (BioLegend, San Diego, CA, USA) was used 1 h before cell stimulation, which was sufficient to deplete IL-10 expression (Supplementary file S1). The anti-CD282 protein specifically produced by clone TL2-1 has been previously reported to cross-react with most mammals, including swine.
RNA Isolation and Microarray Analysis
Total RNA samples for microarray analysis were extracted using the miRNAeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The RNA quantification and quality were confirmed by NanoDrop ND-1000 (Thermo Fisher). Samples were prepared using 2.5 vol of absolute ethanol and 1/10 vol of 3 M sodium acetate. A total of 2 μg RNA from each sample was sent for genome-wide microRNA microarray analysis using μParaflo® microfluidic biochip technology; this service was provided by LC Sciences (Houston, TX, USA). The array consists of 407 pig-specific probes (Sus scrofa) that detect mature miRNA sequences listed in miRBase release 21 (Supplementary file S2). Mature miRNA is the active form of a miRNA, able to associate itself with the RISC (RNA-induced silencing complex) protein complex that facilitates binding to its target. The chip contained multiple internal control probes selected by the service provider, including PUC2PM-20B and PUC2MM-20B. The microarray data can be accessed in the Gene Ontology (GEO) datasets of the National Center for Biotechnology Information (NCBI) (GEO accession GSE132995) and includes data from 3 samples: unstimulated monocytes (GSM3897865), monocytes treated with Bb12 (GSM3897866), and monocytes treated with Bb12 and TLR2-blocking antibody before stimulation (GSM3897867).
RT-qPCR miRNA Validation
RT-qPCR validation confirms that upregulation and downregulation patterns are similar in the miRNAs analyzed on the microarray. RT-qPCR was performed according to the protocol published by Mentzel et al. [10]. For cDNA synthesis, M-MuLV reverse transcriptase (New England Biolabs, Ipswich, MA, USA) was used, and the conditions for reverse transcription reaction were 42 °C, 60 min; 95 °C, 5 min. For qPCR, the conditions were as follows: 50 °C, 2 min; 95 °C, 1 cycle; 95 °C, 15 s; and 60 °C, 1 min, 40 cycles. PCR reactions were performed in duplicate on a QuantStudio 6.0 Flex real-time PCR system (Thermo Fisher) in a 10-μL final volume including 2 μL HOT FIREPol® EvaGreen® qPCR Mix Plus (Solis BioDyne, Tartu, Estonia), 1 μL of each primer at 10 μM, 1 μL of a 1:4 dilution of the cDNA, and 5 μL of DEPC-treated water. The miRNA primers (Table 1) were designed using the publicly available software miRprimer [11]. The amplicon length for measured microRNAs was 51 bp. To select appropriate reference miRNAs, we used RefFinder, a web tool that integrates the currently available major computational programs (geNorm, NormFinder, BestKeeper, and the comparative Delta-Ct method), to compare and rank the tested candidate reference genes [12]. miR-148b-3p was used to normalize the level of miRNA expression since it did not present significant changes to either treatment. Results on miR-148b-3p are shown as the log2 fold change between unstimulated monocytes and Bb12-stimulated monocytes, which confirms that the upregulation and downregulation pattern is similar in the miRNAs analyzed by both tests. The relative miRNA expression level was calculated using the 2–∆∆Ct method, and log2 was used for expressing the fold change.
Statistical Analysis
Eight experimental repeats of each treatment (unstimulated monocytes, Bb12, anti-TLR2 + Bb12) were analyzed by ANOVA to determine differences between treatments. A t-test clustering heat map of statistically significant miRNAs (p < 0.01) was performed by the microarray provider (LC Sciences) to analyze the effect of anti-TLR2 treatment. The GraphPad Prism 6.0 statistical software (GraphPad Software Inc., San Diego, CA, USA) was used to perform multiple t-tests using the Holm–Sidak methods for mean fluorescence intensity (MFI) differences among treatments of selected miRNAs for further analysis with the bioinformatic tools.
Bioinformatic Analysis
miRPath v.3 from DIANA TOOLS [13] was used to investigate the miRNA pathways. miRTarBase 7.0 [14] was used to analyze previously reported validated miRNA–mRNA interactions. Validated interactions were considered only from those reports that showed evidence for miRNA–target interactions (MTIs) where the validation method included either reporter assay, western blot, or qPCR. The Cytoscape software (version 3.7.1) was used to construct a miRNA–gene network. TargetScanHuman 7.2 [15] was used for the search and confirmation of human–pig sequence homology.
Results
miRNA Expression Profile in Response to Bb12
To investigate miRNAs involved in the immune response regulation to Bb12, the miRNA expression profile was analyzed 4 h after stimulation. To focus on TLR2, its blockade was achieved using an anti-TLR2 antibody to analyze the miRNA expression pattern in response to Bb12 without TLR2 participation.
The microarray data analysis was performed according to the diagram presented in Fig. 1. The statistical analysis by ANOVA report showed that expression of 144 miRNAs (144/407) in monocytes (unstimulated monocytes) changed (p < 0.05) in response to the treatments (monocytes treated with Bb12 (Bb12 group) and monocytes treated with Bb12 and TLR2-blocking antibody before stimulation (anti-TLR2 + Bb12 group)); these data are shown in Supplementary file S3. However, the selection criteria for the bioinformatic analysis were to consider only those miRNAs that showed significant changes in their expression and MFI threshold greater than 500 in each sample at a 1% significance level. As shown in Table 2, the statistical analysis by ANOVA indicated that 40 miRNAs (40/407) were differentially expressed in response to the treatments (p < 0.01).
Flowchart of microarray data analysis. Swine monocytes were exposed to Bb12 with or without the addition of a blocking antibody anti-TLR2. Groups were as follows: unstimulated swine monocytes, monocytes stimulated with Bb12 for 4 h, and swine monocytes incubated with anti-TLR2 antibody and Bb12 for 4 h. Microarray was performed, and 40 microRNAs with a MFI > 500 were selected for analysis of molecular interactions (KEGG analysis). The microRNAs were also analyzed for miRNA–mRNA interactions within TLR2 pathway targets (miRTarBase). After this, 15 miRNAs with a reported interaction with TLR2 pathway–related target proteins were selected to analyze with a multiple t test
Integration of Microarray Expression in Response to Bb12 with Experimentally Validated MTIs Databases
Several target genes were found for differentially expressed miRNAs, and all of them were involved in a broad spectrum of cellular, metabolic, physiological, and developmental processes. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using miRPath v.3 (Fig. 2). The heat map displays correlations for 30 miRNAs with available experimental support and their mediated significant pathways (p < 0.05). The statistical significance of interaction is visually represented in varying shades of color (darker colors indicate lower p-values and thus higher significance). We found interactions with more than 100 genes in pathways such as cancer, MAPK (mitogen-activated protein kinases) signaling, proteoglycans in cancer, protein processing in the endoplasmic reticulum, and endocytosis (Supplementary file S4).
Correlation heat map among differentially expressed miRNAs and KEGG pathways (log scaled). The heat map displays statistically significant correlations among 30 differentially expressed miRNAs and their mediated pathways. The color key is on the upper left of the figure; red represents a higher statistical significance whereas yellow indicates a lower statistical significance (p < 0.05). Acronym definitions: ECM, extracellular matrix; HTLV-1, human T-cell lymphotropic virus type 1; MAPK, mitogen-activated protein kinase; PI3K-Akt, phosphatidylinositol 3-kinase (PI3K) and AKT/protein kinase B pathway; FoxO, Forkhead box Class O; TGF, tumor growth factor
We searched for TLR2 pathway–related miRNA–mRNA interactions of the 40 differentially expressed miRNAs previously mentioned at miRTarBase 7.0. Validated interactions were considered only from those reports that showed strong evidence for MTIs where the validation method included either reporter assay, western blot, or qPCR.
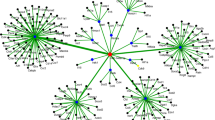
As a result, 15 miRNAs (15/40) had a reported interaction with TLR2 pathway–related target proteins. This information was used to construct a miRNA–gene network. MTIs involve 15 miRNAs and 26 TLR2 pathway–related target proteins (Fig. 3). We used TargetScanHuman 7.2 to ensure interactions obtained in human databases with swine data. Interaction sites of miRNA–mRNA of 18 species were analyzed; of those, 21 mRNA sequences showed homology, 6 did not show homology, and 13 were not found in the TargetScanHuman 7.2 database (Supplementary file S5).
miRNA–mRNA regulatory network in response to Bb12. The clustering network consists of 15 differentially expressed miRNAs and 26 TLR2 pathway–related target proteins. Hexagons represent mRNAs; rhombuses represent miRNAs. The bar-headed lines represent miRNA–mRNA interaction. Acronym definitions: IL-1b, interleukin 1 beta; MyD88, myeloid differentiation primary response 88; MAP2K3, mitogen-activated protein 2 kinase 3; IL-12, interleukin 12; IRAK 1, interleukin receptor–associated kinase 1; TRAF6, TNF receptor–associated factor 6; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; TAB2, TGF-beta-activated kinase 1 and MAP3K7-binding protein 2; JUN, subunit of transcription factor AP-1; FOS, Fos proto-oncogene.
TLR2-Related miRNA Expression Profile in Response to Bb12
Cluster analysis revealed that TLR2 blockade caused changes in miRNA expression as shown in Fig. 4, which represents a clustering heat map of all statistically significant miRNAs (p < 0.01). Focusing on the effect of TLR2 blocking on the 15 miRNAs analyzed, a multiple t-test analysis was performed (Fig. 5). Data were graphed grouping by MFI ranges, considering the differences in the cellular abundance of miRNAs [16]. Those with very low abundance MFI from 1000 to 3000 were grouped in Fig. 5A, low abundance in Fig. 5B (from 3000 to 6000 MFI), high abundance in Fig. 5C (from 6000 to 10,000 MFI), and very high abundance in Fig. 5D (more than 10,000 MFI). Among miRNAs upregulated by Bb12 treatment included were miR-29b-3p, miR-30d-5p, miR-181a-5p (Fig. 5B), miR-15a-5p (Fig. 5C), miR-16-5p, and miR-26a-5p (Fig. 5D), whereas miR-221-3p (Fig. 5A), miR-19b-3p (Fig. 5B), miR-23a-5p (Fig. 5C), and miR-21-5p (Fig. 5D) were downregulated (p < 0.01). Expression of let-7c-5p, miR-146b-5p, miR-155-5p (Fig. 5A), let-7f-5p (Fig. 5C), and miR-150-5p (Fig. 5D) increased only when TLR2 was blocked (p < 0.01).
t-test clustering heat map of statistically significant miRNAs (p < 0.01) due to the effect of anti-TLR2 treatment. A t-test was performed for the expression levels of each miRNA comparing the group of eight experimental repeats with only Bb12 to the group of eight experimental repeats with anti-TLR2 + Bb12.
MFI changes in response to Bb12 with or without the TLR2 blockade. Graphs show miRNAs with TLR2-related target proteins that presented interaction evidence. Data were graphed grouping by MFI ranges, considering the differences in the cellular abundance of miRNAs, from low to high intensity. A, very low; B, low; C, high; and D, very high abundance. Statistical significance was determined using multiple t-test Holm–Sidak methods. Data are shown as mean ± SD, n = 8 experimental repeats. Different letters indicate a significant difference between groups (p < 0.01)
RT-qPCR miRNA Validation
To validate microarray results, the expression level of five miRNAs was confirmed by RT-qPCR with the same samples. The trend for let-7 members was maintained as downregulation, showing a log2 fold change of − 0.58 for let-7c-5p and − 0.52 for let-7d-5p, and neither showed significant changes in response to Bb12 according to the t-test (Supplementary file S6). MiR-20a-5p and miR-21-5p were significantly downregulated (p < 0.05) on the microarray with a log2 fold change of − 0.17 and − 0.16, respectively, confirmed by RT-qPCR with a fold change of − 0.29 for miR-20a-5p and − 0.26 for miR-21-5p. The miR-30a-5p expression indicated upregulation; however, there were no significant changes according to the microarray t-test results (Fig. 6).
Discussion
Bb12 modulates the immune system, affecting gene expression and cytokine production [17] due to interaction with receptors like TLR2 [18]. Probiotic bacteria can promote tolerant immune responses to gain allowance for the establishment in their environment while offering different benefits to the host. This is a complicated process and needs an accurate response; therefore, TLR-activated pathways are strictly regulated. During the last decade, microRNAs have been demonstrated to be one of the main molecules implicated in the post-transcriptional regulation of immune responses.
Immune responses toward probiotics are strain-dependent, which could be attributed to differences in the expression of regulators, such as miRNAs. In this study, miR-15a-5p, miR-29b-3p, miR-30d-5p, and miR-181a-5p were upregulated by Bb12, and their expression was shown to be TLR2-dependent since it was abrogated upon TLR2 blocking. The Bb12 immunomodulation mechanism initiates with TLR2 recognition of PGN or LTA present in the bacterial membrane, which in turn promotes the activation of transcription factors like AP-1 and NF-κB [19, 20]. miR-15a-5p has been reported as a tumor suppressor by inhibiting the expression of Bcl-2 (B-cell lymphoma 2), an important anti-apoptotic protein. The Bcl-2 expression can be induced by NF-κB [21]; therefore, these three molecules could represent a regulation network for tolerance induction. miR-29b-3p has been reported to bind c-Fos (Finkel–Biskis–Jinkins murine osteogenic sarcoma virus human homolog proto-oncogene) [22], which is the main component of transcription factor AP-1; hence, the high expression of miR-29b-3p could regulate inflammatory responses by inhibiting precursors of AP-1. This miRNA has also been reported as a potential diagnostic biomarker of pathogenesis since its downregulation can increase NF-κB activation when stimulated by cytokines, resulting in enhanced inflammation [23].
MAPK proteins also have an impact on NF-κB and AP-1 activation, and consequently on cytokine production. Our results indicate an upregulation of miR-30d and miR-181a, which have been reported to regulate MAPK4K4, MAPK1, and MAP2K1. The overexpression of miR-30d protects cells from TNF-α (tumor necrosis factor-alpha) suppression via downregulation of MAP4K4 in pancreatic β-cells [24]. The miR-181 family has been reported to be an effector in the inflammatory response [25] and functions as a bona fide tumor suppressor in salivary adenoid cystic carcinoma [26]. They are also involved in c-Fos regulation and therefore reported as a potentially novel mechanism contributing to immune homeostasis in atherosclerosis [27]. Both miR-30d and miR-181a are involved in the homeostasis establishment process and could be important regulators of tolerant responses toward probiotics.
TLR2 blockade shows how recognition of Bb12 is also mediated by other receptors since the production of cytokines was also present during the blockade. MiR-16-5p and miR-26a-5p had an increased expression in swine monocytes when TLR2 was blocked, demonstrating that their expression does not depend on TLR2–bacteria interaction. MiR-16-5p and miR-26a-5p are transcribed as a cluster, and their presence was associated with a decreased expression of ERK (extracellular signal-regulated kinase), a mediator kinase between bacterial recognition and transcription factors such as AP-1. The miR-15a-5p decrease is reported to be related to cancer processes [28], and miR-16-5p is a potential biomarker of different types of cancers and since it has a stable expression in breast cancer tissue and gastric cancer, it is commonly used as an endogenous control in MTIs [29, 30]. According to the bioinformatic analysis of MTIs, miR-26a-5p can regulate the expression of pro-inflammatory cytokines IFN-α, IL-6, and IL-12. Contrarily, miR-26a-5p expression decreases in various tumor types, functioning as a tumor suppressor [31].
Let-7c-5p and let-7f-5p had an increased expression in monocytes stimulated with Bb12 during the TLR2 blockade. Members of the let-7 family have been described as a pro-inflammatory miRNA with immunomodulatory function [32,33,34]. Particularly, let-7-5p participates in cancer processes, as a regulator of innate immunity networks and in response to TLR4 activation by lipopolysaccharide [35,36,37,38,39]. Other miRNAs which expression augmented when TLR2 was blocked included miR-146b-5p, miR-155-5p, and miR-150-5p. According to the interaction analysis, miR-146b-5p could regulate key proteins involved in signaling pathways of the immune response: IRAK1 (interleukin receptor–associated kinase 1), TRAF6 (TNF receptor–associated factor 6), NF-κB1, and IL-6; therefore, an increase of miR-146b-5p due to TLR2 absence could affect these key proteins. Reports show that expression of miR-146b-5p increases upon monocyte differentiation into dendritic cells (DCs), while its silencing prevents DC apoptosis [40].
Zeuthen et al. [41] showed that bifidobacteria inhibit TNF-α and IL-6 production in bone marrow–derived DCs from TLR2−/− mice. In this same article, bifidobacteria induce much higher levels of IL-12 and lower IL-10 levels in DCs that are TLR2−/− when compared with wild-type DCs [41]. In our study, an increase in let-7c-5p expression was observed using a TLR2 blockade, which gives a glimpse of the mechanisms involved in the decrease of IL-10 expression observed upon TLR2 blockade in swine monocytes [8]. An increase in let-7c-5p expression could be associated with an IL-10 decrease since let-7c-5p is reported to regulate IL-10 expression [42]. These results suggest an anti-inflammatory role of the receptor, as it has been previously demonstrated by Peres et al. [43] and Quero et al. [44] and its critical role in the immunomodulatory function of Bb12.
Bb12 stimulation downregulated miR-19b-3p and miR-23a-5p; nonetheless, their expressions were also downregulated when TLR2 was blocked. Both miRNAs can regulate the transcript of TLR2 [45, 46], which could indicate the participation of other pattern recognition receptors acting redundantly in the recognition of Bb12, downregulating those miRNAs. For instance, maltose-binding proteins (MBPs) are one of the many molecules in the bifidobacterial cell wall that can be recognized by TLR4 and induce maturation of DCs [47, 48]. In macrophages, they promote M1 polarization and upregulate TLR2, MyD88, and IκB-α mRNA expression, and these effects can be largely abrogated by a pretreatment blockade of TLR2 or TLR4 [49]. DC-SIGN is a C-type lectin receptor that participates in DC maturation activating transcription factor PU.1, which is a target for miR-155 [50]. Recognition of probiotics by DC-SIGN primes monocyte-derived DCs to drive the development of Treg cells, which increase the levels of IL-10. By doing so, IL-10 production can stall inflammatory responses and inhibit the proliferation of bystander T cells [51].
Intracellularly, CpG-oligodeoxynucleotide recognition by TLR9 causes an upregulation of miR-146a, which targets Notch1 and represses IL-12p70 production in mice DCs [52]. Live bacteria were used in our experiment; therefore, we cannot dismiss the possibility that bacterial CpG sequences are being recognized and contribute to miRNA expression and immune response modulation. PGN in probiotic bacteria is recognized by the intracellular receptor nucleotide-binding oligomerization domain-containing protein 2 (NOD2) [53, 54], which can positively or negatively regulate NF-κB/MAPK-dependent pro-inflammatory cytokines [55, 56]. Although it has been demonstrated that mutations in NOD2 are linked to aberrant immune responses, activation of the TLR2‐NOD2‐RIPK2 pathway affects IL-10 secretion and consequently the balance between pro‐ and anti‐inflammatory responses to Gram‐positive bacteria [57].
Lastly, miRNAs can also contribute as physiological ligands for TLRs, a function that is independent of their conventional role in post-transcriptional gene regulation [58]. Exosomes containing miRNAs have been reported to be transferred from cell to cell and eventually reach TLR-containing endosomes [59]. TLR7 and TLR8 can recognize miRNAs, and these binding miRNAs function as agonists of these single-stranded RNA-binding TLRs, which can lead to NF-kB signaling activation and secretion of interleukin IL-6 and TNF-ɑ, which in turn promote cancer cell growth and metastasis [59, 60].
In this study, miR-15a-5p, miR-29b-3p, miR-30d-5p, and miR-181a-5p were upregulated by Bb12 and their expression was TLR2-dependent. These miRNAs inhibit proteins that participate in inflammatory responses, therefore representing important candidates in the induction of tolerance toward probiotics. Let-7c-5p, let-7f-5p, miR-16-5p, miR-26a-5p, miR-146b-5p, miR-150-5p, and miR-155-5p had an increased expression with TLR2 blockade, demonstrating that their expression does not depend on TLR2–bacteria interaction. The increase of let-7c-5p expression is particularly interesting since it is capable of regulating IL-10, and a previous publication of our group showed that IL-10 expression is affected by TLR2 blocking. This could imply a regulatory network, but more experiments are required to clarify the connection between TLR2, IL-10, and let-7c-5p. Inhibition of TLR2 abrogated its participation in the development of an anti-inflammatory response but demonstrated that Bb12 recognition is complemented by other receptors since there were still changes in the microtranscriptome. These changes have potential effects in downstream proteins such as MyD88, TRAF6, IRAK1, TAB2 (tumor growth factor-beta activated kinase 1), and MAPK members; transcription factors such as NF-κB and AP-1; and cytokines such as IL-1A, IL-2, IL-6, IL-10, IL-12, and TNF-α.
In conclusion, TLR2 is pivotal for Bb12 immunomodulation because of its anti-inflammatory function. This could be a cross-linked mechanism between the TLR2 pathway and other cell routes activated by bifidobacteria recognition. In this mechanism miRNAs are involved; they could be induced through the participation of different pattern recognition receptors, and their expression regulates the TLR2 pathway.
Probiotics have proved to offer several benefits, and their recognition can induce a tolerant immune response, unlike when pathogens are recognized. Nevertheless, the molecular mechanisms within this complex process are still being elucidated. Post-transcriptional regulation by miRNAs in cellular signaling impacts several key proteins, like receptors, signaling pathway members, transcription factors, and cytokines. Accurate regulation of signaling pathways on behalf of the recognition of microbes is fundamental for efficient and differential responses between probiotics and pathogens. These differential responses are also related to their capability of inducing changes in the microtranscriptome. Knowledge generated about how probiotic recognition induces changes in the microtranscriptome will contribute to a deeper understanding of their immunomodulatory mechanisms.
Data Availability
The datasets analyzed during the current study are available in the Gene Ontology (GEO) datasets of the National Center for Biotechnology Information (NCBI) (GEO accession GSE132995).
References
Jungersen M, Wind A, Johansen E, Christensen JE, Stuer-Lauridsen B, Eskesen D (2014) The science behind the probiotic strain Bifidobacterium animalis subsp. lactis BB-12®. Microorganisms 2:92–110. https://doi.org/10.3390/microorganisms2020092
López P, Gueimonde M, Margolles A, Suárez A (2010) Distinct Bifidobacterium strains drive different immune responses in vitro. Int J Food Microbiol 138:157–165. https://doi.org/10.1016/j.ijfoodmicro.2009.12.023
Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. https://doi.org/10.1038/ni.1863
Valeriano VDV, Balolong MP, Kang DK (2017) Probiotic roles of Lactobacillus sp. in swine: insights from gut microbiota. J Appl Micro 122:554–567. https://doi.org/10.1111/jam.13364
Barba-Vidal E, Castillejos L, López-Colom P, Rivero Urgell M, Moreno Muñoz JA, Martín-Orúe SM (2017) Evaluation of the probiotic strain Bifidobacterium longum subsp. infantis CECT 7210 capacities to improve health status and fight digestive pathogens in a piglet model. Front Microbiol 8:533. https://doi.org/10.3389/fmicb.2017.00533
Hvistendahl M (2012) Pigs as stand-ins for microbiome studies. Science 336:1250. https://doi.org/10.1126/science.336.6086.1250
Walters EM, Prather RS (2013) Advancing swine models for human health and diseases. Mo Med 110:212–215
Arenas-Padilla M, Duarte-Gutiérrez JL, Mata-Haro V (2018) Bifidobacterium animalis ssp. lactis Bb12 induces IL-10 through cell membrane-associated components via TLR2 in swine. J Appl Microbiol 125:1881–1889. https://doi.org/10.1111/jam.14069
Pasquinelli AE (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13:271–282. https://doi.org/10.1038/nrg3162
Mentzel CMJ, Skovgaard K, Cordoba S, Herrera J, Busk PK, Cirera S (2014) Wet-lab tested MicroRNA assays for qPCR studies with SYBR® green and DNA primers in pig tissues. MicroRNA 3:174–188. https://doi.org/10.2174/2211536604666141226194231
Busk PK (2014) A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics 15:29. https://doi.org/10.1186/1471-2105-15-29
Xie F, Xiao P, Chen D, Xu L, Zhang B (2012) miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 80:75–84. https://doi.org/10.1007/s11103-012-9885-2
Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG (2015) DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 43:W460–W466. https://doi.org/10.1093/nar/gkv403
Chou C-H, Shrestha S, Yang C-D, Chang N-W, Lin Y-L, Liao K-W, Huang W-C, Sun T-H, Tu S-J, Lee W-H (2017) miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 46:D296–D302. https://doi.org/10.1093/nar/gkx1067
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005. https://doi.org/10.7554/eLife.05005
Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E (2016) Distribution of miRNA expression across human tissues. Nucleic Acids Res 44:3865–3877. https://doi.org/10.1093/nar/gkw116
Zarrati M, Shidfar F, Nourijelyani K, Mofid V, Hosseinzadeh-Attar MJ, Bidad K, Najafi F, Gheflati Z, Chamari M, Salehi E, (2013) Lactobacillus acidophilus La5, Bifidobacterium BB12, and Lactobacillus casei DN001 modulate gene expression of subset specific transcription factors and cytokines in peripheral blood mononuclear cells of obese and overweight people. Biofactors 39:633–643. https://doi.org/10.1002/biof.1128
Meng H, Ba Z, Lee Y, Peng J, Lin J, Fleming JA, Furumoto EJ, Roberts RF, Kris-Etherton PM, Rogers CJ (2017) Consumption of Bifidobacterium animalis subsp. lactis BB-12 in yogurt reduced expression of TLR-2 on peripheral blood-derived monocytes and pro-inflammatory cytokine secretion in young adults. Eur J Nutr 56:649–661. https://doi.org/10.1007/s00394-015-1109-5
González-Rascón A, Mata-Haro V (2015) MicroRNAs: regulators of TLR2-mediated probiotic immune responses. MicroRNA 4:168–174. https://doi.org/10.2174/2211536605666151208123209
Tomosada Y, Villena J, Murata K, Chiba E, Shimazu T, Aso H, Iwabuchi N, Xiao J-z, Saito T, Kitazawa H (2013) Immunoregulatory effect of bifidobacteria strains in porcine intestinal epithelial cells through modulation of ubiquitin-editing enzyme A20 expression. PLOS ONE 8:e59259. https://doi.org/10.1371/journal.pone.0059259
Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, Wang H, Sun H, Volinia S, Alder H, Calin GA, Liu CG, Andreeff M, Croce CM (2007) MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene 26:4148–4157. https://doi.org/10.1038/sj.onc.1210186
Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E, Botta C, Paolino FM, Del Giudice T, Iuliano E (2013) miR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol 228:1506–1515. https://doi.org/10.1002/jcp.24306
Xing L-N, Wang H, Yin P-H, Liu Y-J, Chi Y-F, Wang Y-M, Peng W (2014) Reduced miR-29b-3p expression up-regulate CDK6 and contributes to IgA nephropathy. Int J Clin Exp Med 15:5275–5281
Zhao X, Mohan R, Özcan S, Tang X (2012) MicroRNA-30d induces insulin transcription factor MafA and insulin production by targeting mitogen-activated protein 4 kinase 4 (MAP4K4) in pancreatic β-cells. J Biol Chem 287:31155–31164. https://doi.org/10.1074/jbc.M112.362632
Huang X, Schwind S, Santhanam R, Eisfeld A-K, Chiang C-l, Lankenau M, Yu B, Hoellerbauer P, Jin Y, Tarighat SS (2016) Targeting the RAS/MAPK pathway with miR-181a in acute myeloid leukemia. Oncotarget 7:59273–59286. https://doi.org/10.18632/oncotarget.11150
He Q, Zhou X, Li S, Jin Y, Chen Z, Chen D, Cai Y, Liu Z, Zhao T, Wang A (2013) MicroRNA-181a suppresses salivary adenoid cystic carcinoma metastasis by targeting MAPK–Snai2 pathway. Biochim Biophys Acta 1830:5258–5266. https://doi.org/10.1016/j.bbagen.2013.07.028
Wu C, Gong Y, Yuan J, Zhang W, Zhao G, Li H, Sun A, Zou Y, Ge J (2012) microRNA-181a represses ox-LDL-stimulated inflammatory response in dendritic cell by targeting c-Fos. J Lipid Res 53:2355–2363. https://doi.org/10.1194/jlr.M028878
Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’urso L, Pagliuca A, Biffoni M, Labbaye C, (2008) The miR-15a–miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 14:1271–1277. https://doi.org/10.1038/nm.1880
Rinnerthaler G, Hackl H, Gampenrieder SP, Hamacher F, Hufnagl C, Hauser-Kronberger C, Zehentmayr F, Fastner G, Sedlmayer F, Mlineritsch B, Greil R (2016) miR-16-5p is a stably-expressed housekeeping microRNA in breast cancer tissues from primary tumors and from metastatic sites. Int J Mol Sci 17:156. https://doi.org/10.3390/ijms17020156
Zhang W, Shen X, Xie L, Chu M, Ma Y (2015) MicroRNA-181b regulates endotoxin tolerance by targeting IL-6 in macrophage RAW264.7 cells. J Inflamm (Lond) 12:18. https://doi.org/10.1186/s12950-015-0061-8
Gao J, Liu Q-G (2011) The role of miR-26 in tumors and normal tissues. Oncol Lett 2:1019–1023. https://doi.org/10.3892/ol.2011.413
Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, Gupta P, Raffetseder J, Lerm M, Ghosh Z (2015) MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe 17:345–356. https://doi.org/10.1016/j.chom.2015.01.007
Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA (2010) Pro-inflammatory role for let-7 microRNAs in experimental asthma. J Biol Chem 285:30139–30149. https://doi.org/10.1074/jbc.M110.145698
Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J (2011) Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J 30:1977–1989. https://doi.org/10.1038/emboj.2011.94
Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6:259–269. https://doi.org/10.1038/nrc1840
Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11:441–450. https://doi.org/10.1016/j.devcel.2006.09.009
Ren Z, Ambros VR (2015) Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc Natl Acad Sci 112:E2366–E2375. https://doi.org/10.1073/pnas.1422858112
Shi X-B, Tepper CG, deVere White RW (2008) Cancerous miRNAs and their regulation. Cell Cycle 7:1529–1538. https://doi.org/10.4161/cc.7.11.5977
Zhang M, Liu F, Jia H, Zhang Q, Yin L, Liu W, Li H, Yu B, Wu J (2011) Inhibition of microRNA let-7i depresses maturation and functional state of dendritic cells in response to lipopolysaccharide stimulation via targeting suppressor of cytokine signaling 1. J Immunol 187:1674–1683. https://doi.org/10.4049/jimmunol.1001937
Park H, Huang X, Lu C, Cairo MS, Zhou X (2015) MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J Biol Chem 290:2831–2841. https://doi.org/10.1074/jbc.M114.591420
Zeuthen LH, Fink LN, Frøkiær H (2008) Toll-like receptor 2 and nucleotide-binding oligomerization domain-2 play divergent roles in the recognition of gut-derived lactobacilli and bifidobacteria in dendritic cells. Immunology 124:489–502. https://doi.org/10.1111/j.1365-2567.2007.02800.x
Jiang L, Cheng Z, Qiu S, Que Z, Bao W, Jiang C, Zou F, Liu P, Liu J (2012) Altered let-7 expression in Myasthenia gravis and let-7c mediated regulation of IL-10 by directly targeting IL-10 in Jurkat cells. Int Immunopharmacol 4:217–223. https://doi.org/10.1016/j.intimp.2012.07.003
Peres A, Stegen C, Cousineau B, Desrosiers M, Madrenas J (2013) The pro-inflammatory and anti-inflammatory TLR2 responses to Staphylococcus aureus can be uncoupled (P1242). J Immunol 190(138):19
Quero L, Hanser E, Manigold T, Tiaden AN, Kyburz D (2017) TLR2 stimulation impairs anti-inflammatory activity of M2-like macrophages, generating a chimeric M1/M2 phenotype. Arthritis Res Ther 19:245. https://doi.org/10.1186/s13075-017-1447-1
Gu X, Gao Y, Mu D-G, Fu E-Q (2017) MiR-23a-5p modulates mycobacterial survival and autophagy during Mycobacterium tuberculosis infection through TLR2/MyD88/NF-κB pathway by targeting TLR2. Exp Cell Res 354:71–77. https://doi.org/10.1016/j.yexcr.2017.03.039
Philippe L, Alsaleh G, Suffert G, Meyer A, Georgel P, Sibilia J, Wachsmann D, Pfeffer S (2012) TLR2 expression is regulated by microRNA miR-19 in rheumatoid fibroblast-like synoviocytes. J Immunol 184:454–461. https://doi.org/10.4049/jimmunol.1102348
Fernandez S, Palmer DR, Simmons M, Sun P, Bisbing J, McClain S, Mani S, Burgess T, Gunther V, Sun W (2007) Potential role for toll-like receptor 4 in mediating Escherichia coli maltose-binding protein activation of dendritic cells. Infect Immun 75:1359–1363. https://doi.org/10.1128/IAI.00486-06
Gilad O, Svensson B, Viborg AH, Stuer-Lauridsen B, Jacobsen S (2011) The extracellular proteome of Bifidobacterium animalis subsp. lactis BB-12 reveals proteins with putative roles in probiotic effects. Proteomics 11:2503–2514. https://doi.org/10.1002/pmic.201000716
Ni W, Zhang Q, Liu G, Wang F, Yuan H, Guo Y, Zhang X, Xie F, Li Q, Tai G (2014) Escherichia coli maltose-binding protein activates mouse peritoneal macrophages and induces M1 polarization via TLR2/4 in vivo and in vitro. Int Immunopharmacol 21:171–180. https://doi.org/10.1016/j.intimp.2014.04.025
Martinez-Nunez RT, Louafi F, Friedmann PS, Sanchez-Elsner T (2009) MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN). J Biol Chem 284:16334–16342. https://doi.org/10.1074/jbc.M109.011601
Smits HH, Engering A, van der Kleij D, de Jong EC, Schipper K, van Capel TMM, Zaat BAJ, Yazdanbakhsh M, Wierenga EA, van Kooyk Y, Kapsenberg ML (2005) Selective probiotic bacteria induce IL-10–producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell–specific intercellular adhesion molecule 3–grabbing nonintegrin. J Allergy Clin Immunol 115:1260–1267. https://doi.org/10.1016/j.jaci.2005.03.036
Bai Y, Qian C, Qian L, Ma F, Hou J, Chen Y, Wang Q, Cao X (2012) Integrin CD11b negatively regulates TLR9-triggered dendritic cell cross-priming by upregulating microRNA-146a. J Immunol 188:5293–5302. https://doi.org/10.4049/jimmunol.1102371
Habil N, Al-Murrani W, Beal J, Foey A (2011) Probiotic bacterial strains differentially modulate macrophage cytokine production in a strain-dependent and cell subset-specific manner. Benef Microbes 2:283–293. https://doi.org/10.3920/BM2011.0027
Hasegawa M, Yang K, Hashimoto M, Park J-H, Kim Y-G, Fujimoto Y, Nuñez G, Fukase K, Inohara N (2006) Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem 289:29054–29063. https://doi.org/10.1074/jbc.M602638200
Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278:8869–8872. https://doi.org/10.1074/jbc.C200651200
Rosenstiel P, Huse K, Till A, Hampe J, Hellmig S, Sina C, Billmann S, von Kampen O, Waetzig GH, Platzer M (2006) A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc Natl Acad Sci 103:3280–3285. https://doi.org/10.1073/pnas.0505423103
Moreira LO, El Kasmi KC, Smith AM, Finkelstein D, Fillon S, Kim YG, Núñez G, Tuomanen E, Murray PJ (2008) The TLR2-MyD88-NOD2-RIPK2 signalling axis regulates a balanced pro-inflammatory and IL-10-mediated anti-inflammatory cytokine response to Gram-positive cell walls. Cell Microbiol 10:2067–2077. https://doi.org/10.1111/j.1462-5822.2008.01189.x
Chen X, Liang H, Zhang J, Zen K, Zhang CY (2013) microRNAs are ligands of Toll-like receptors. RNA 19:737–739. https://doi.org/10.1261/rna.036319.112
Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM (2012) MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci 109:E2110–E2116. https://doi.org/10.1073/pnas.1209414109
Winkler CW, Taylor KG, Peterson KE (2014) Location is everything: let-7b microRNA and TLR7 signaling results in a painful TRP. Sci Signal 7:pe14. https://doi.org/10.1126/scisignal.2005407
Funding
This research was funded by CONACYT grant number 267462. MAP was granted a Ph.D. research scholarship by CONACYT.
Author information
Authors and Affiliations
Contributions
Conceptualization, M.A.P. and V.M.H.; writing—original draft preparation, M.A.P.; writing—review and editing, A.H.M., A.M.C., J. H., and A.G.R.; funding acquisition, J. H. and V.M.H.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Arenas-Padilla, M., González-Rascón, A., Hernández-Mendoza, A. et al. Immunomodulation by Bifidobacterium animalis subsp. lactis Bb12: Integrative Analysis of miRNA Expression and TLR2 Pathway–Related Target Proteins in Swine Monocytes. Probiotics & Antimicro. Prot. 14, 510–522 (2022). https://doi.org/10.1007/s12602-021-09816-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-021-09816-1