Abstract
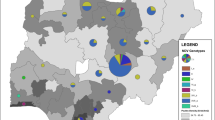
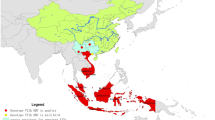
Following recent Newcastle disease virus (NDV) outbreaks in Iranian poultry farms which were mostly associated with lesions of the avian gastrointestinal tract, it was speculated that the scale of the outbreaks could be attributed in part to co-circulating infectious agents or a new NDV genotype/subgenotype. This speculation was due to the isolation of a few 5th panzootic subgenotype VII.2 viruses from Iranian poultry farms in 2017. Samples from different species of commercial and domestic birds were collected from different provinces of Iran, 19 of which were selected for the current study. Phylogenetic analyses showed that the recent outbreaks have been caused by only one agent, i.e. the distinctive NDV subgenotype VII.1.1 (previously known VIIl) viruses that seem to be circulating predominantly in Iran, but have also been sporadically reported from Iraq among neighbouring countries. At most, 96.3–96.7% BLAST identity to non-Iranian VII.1.1 isolates was observed. Genetic distance values of <1% were indicative of high similarity between the isolates, but the values were approaximately 2% when the current isolates were compared to the earliest recorded Iranian VII.1.1 viruses isolated in 2010. Using Bayesian analysis, annual mutation rates of 1.7156E-3 (strict) and 1.9902E-3 (relaxed) over 11 years were obtained. In addition, we report that our laboratories have not detected any genotype XIII strains since 2011. Following up on previous reports, we concluded that currently, and except in Columbiforms, subgenotype VII.1.1 may likely be the predominant subgenotype in many bird species in Iran despite the subgenotype VII.2 being predominant in neighbouring countries.


Similar content being viewed by others
Change history
14 August 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00284-021-02619-1
References
Walker PJ, Siddell SG, Lefkowitz EJ et al (2019) Changes to virus taxonomy and the international code of virus classification and nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch Virol 164:2417–2429. https://doi.org/10.1007/s00705-019-04306-w
Amarasinghe GK, Ayllón MA, Bào Y et al (2019) Taxonomy of the order Mononegavirales: update 2019. Arch Virol 164:1967–1980. https://doi.org/10.1007/s00705-019-04247-4
Yusoff K, Tan WS (2001) Newcastle disease virus: macromolecules and opportunities. Avian Pathol 30:439–455. https://doi.org/10.1080/03079450120078626
Sohrab V (1973) Newcastle disease in Iran. Bull Off Int Epizoot 79:565–569
Fallah Mehrabadi MH, Ghafouri SA, Shoushtari A et al (2020) Effectiveness of thermostable vaccine for newcastle disease produced by the Razi institute on backyard poultry in Iran during 2015. Arch Razi Inst 75:1–7
Allahyari E, Allymehr M, Molouki A et al (2020) Molecular characterisation and phylogenetic study of the fusion gene of Newcastle disease viruses isolated from broiler farms of Iran in 2018–2019. Bulg J Vet Med. https://doi.org/10.15547/bjvm.2020-0041
Molouki A, Mehrabadi MHF, Bashashati M et al (2019) NDV subgenotype VII(L) is currently circulating in commercial broiler farms of Iran, 2017–2018. Trop Anim Health Prod 51:1247–1252. https://doi.org/10.1007/s11250-019-01817-1
OIE (2012) Newcastle disease. In: Manual of diagnostic tests and vaccines for terrestrial animals, vol 1, 7th edn. World Organisation for Animal Health, Paris, France, pp. 555–573
Dimitrov KM, Abolnik C, Afonso CL et al (2019) Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect Genet Evol 74:103917. https://doi.org/10.1016/j.meegid.2019.103917
Dimitrov KM, Ramey AM, Qiu X et al (2016) Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus). Infect Genet Evol 39:22–34. https://doi.org/10.1016/j.meegid.2016.01.008
Diel DG, da Silva LHA, Liu H et al (2012) Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect Genet Evol 12:1770–1779. https://doi.org/10.1016/j.meegid.2012.07.012
Rezaei Far A, Peighambari SM, Pourbakhsh SA et al (2017) Co-circulation of genetically distinct groups of avian paramyxovirus type 1 in pigeon Newcastle disease in Iran. Avian Pathol 46:36–43. https://doi.org/10.1080/03079457.2016.1203068
Mohammadnia Afrouzi S (2019) Annual statistics of the Iranian Ministry of Agriculture - 1398. Ministry of Agriculture, Tehran
Miller PJ, Haddas R, Simanov L et al (2015) Identification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic features. Infect Genet Evol 29:216–229. https://doi.org/10.1016/j.meegid.2014.10.032
Wajid A, Dimitrov KM, Wasim M et al (2017) Repeated isolation of virulent Newcastle disease viruses in poultry and captive non-poultry avian species in Pakistan from 2011 to 2016. Prev Vet Med 142:1–6. https://doi.org/10.1016/j.prevetmed.2017.04.010
Hirschinger J, Munoz MC, Hingrat Y et al (2020) Exposure to and circulation of avian influenza and newcastle disease viruses in peridomestic wild birds in the United Arab Emirates. J Wildl Dis 56:437–442. https://doi.org/10.7589/2019-06-164
Alsahami AA, Ideris A, Omar A et al (2018) Isolation, identification and molecular characterization of Newcastle disease viruses in vaccinated chickens from commercial farms in the Sultanate of Oman. Int J Vet Sci Med 6:248–252. https://doi.org/10.1016/j.ijvsm.2018.08.007
Ghalyanchilangeroudi A, Hosseini H, Jabbarifakhr M et al (2018) Emergence of a virulent genotype VIIi of Newcastle disease virus in Iran. Avian Pathol 47:509–519. https://doi.org/10.1080/03079457.2018.1495313
Soltani M, Peighambari SM, Pourbakhsh SA et al (2019) Phylogenetic study of two Newcastle disease virus (NDV) isolates obtained from poultry flocks in Isfahan Province in 1999 based on haemagglutinin-neuraminidase (HN) gene sequencing. Iran J Vet Res 74:396–407. https://doi.org/10.22059/jvr.2019.225699.2576
Ebrahimi MM, Shahsavandi S, Moazenijula G, Shamsara M (2012) Phylogeny and evolution of Newcastle disease virus genotypes isolated in Asia during 2008–2011. Virus Genes 45:63–68. https://doi.org/10.1007/s11262-012-0738-5
Samadi S, Kianizadeh M, Najafi MF et al (2014) Molecular characterization and phylogenetic study of velogenic Newcastle disease virus isolates in Iran. Virus Genes 48:290–295. https://doi.org/10.1007/s11262-013-1015-y
Hoffmann E, Stech J, Guan Y et al (2002) Universal primer set for the full-length amplification of all Influenza A viruses. Arch Virol 146:2275–2289. https://doi.org/10.1007/s007050170002
Capua I, Minta Z, Karpinska E et al (1999) Co-circulation of four types of infectious bronchitis virus (793/B, 624/I, B1648 and Massachusetts). Avian Pathol 28:587–592. https://doi.org/10.1080/03079459994380
Kumar S, Stecher G, Li M et al (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Bouckaert R, Vaughan TG, Barido-Sottani J et al (2019) BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol 15:e1006650
Hicks JT, Dimitrov KM, Afonso CL et al (2019) Global phylodynamic analysis of avian paramyxovirus-1 provides evidence of inter-host transmission and intercontinental spatial diffusion. BMC Evol Biol 19:108. https://doi.org/10.1186/s12862-019-1431-2
Rambaut A, Drummond AJ, Xie D et al (2018) Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol 67:901–904. https://doi.org/10.1093/sysbio/syy032
Steensels M, Van Borm S, Mertens I et al (2020) Molecular and virological characterization of the first poultry outbreaks of Genotype VII.2 velogenic avian orthoavulavirus type 1 (NDV) in North-West Europe, BeNeLux, 2018. Transbound Emerg Dis. https://doi.org/10.1111/tbed.13863
Mayahi V, Esmaelizad M (2017) Molecular evolution and epidemiological links study of Newcastle disease virus isolates from 1995 to 2016 in Iran. Arch Virol 162:3727–3743. https://doi.org/10.1007/s00705-017-3536-5
Dimitrov KM, Lee D-H, Williams-Coplin D et al (2016) Newcastle disease viruses causing recent outbreaks worldwide show unexpectedly high genetic similarity to historical virulent isolates from the 1940s. J Clin Microbiol 54:1228–1235. https://doi.org/10.1128/JCM.03044-15
Miller PJ, Kim LM, Ip HS, Afonso CL (2009) Evolutionary dynamics of Newcastle disease virus. Virology 391:64–72. https://doi.org/10.1016/j.virol.2009.05.033
Tavassoli A (1971) Immune response of chickens to four lentogenic strains of Newcastle disease virus propagated in lamb kidney cell culture. Arch Inst Razi 23:129–135
Tavassoli A (1973) Studies on the immunogenic properties of three lentogenic strains of Newcastle disease virus. Arch Inst Razi 25:79–88
Bozorgmehri-Fard MH, Keyvanfar H (1979) Isolation of Newcastle disease virus from teals (Anas crecca) in Iran. J Wildl Dis 15:335–337. https://doi.org/10.7589/0090-3558-15.2.335
Almubarak AIA (2019) Molecular and biological characterization of some circulating strains of Newcastle disease virus in broiler chickens from Eastern Saudi Arabia in 2012–2014. Vet World 12:1668–1676. https://doi.org/10.14202/vetworld.2019.1668-1676
Al-Zuhariy M (2016) Isolation and identification of the Newcastle disease virus from field outbreaks in broiler and layer flocks in Iraq. Iraqi J Vet Med 41:23–27. https://doi.org/10.30539/iraqijvm.v41i1.73
Lomniczi B, Wehmann E, Herczeg J et al (1998) Newcastle disease outbreaks in recent years in Western Europe were caused by an old (VI) and a novel genotype (VII). Arch Virol 143:49–64. https://doi.org/10.1007/s007050050267
Yang C-Y, Shieh HK, Lin Y-L, Chang P-C (1999) Newcastle disease virus isolated from recent outbreaks in Taiwan phylogenetically related to viruses (genotype vii) from recent outbreaks in Western Europe. Avian Dis 43:125–130. https://doi.org/10.2307/1592771
Farkas T, Székely É, Belák S, Kiss I (2009) Real-time PCR-based pathotyping of newcastle disease virus by use of TaqMan minor groove binder probes. J Clin Microbiol 47:2114–2123. https://doi.org/10.1128/JCM.01652-08
Soltani M, Peighambari SM, Pourbakhsh SA et al (2019) Molecular characterization of haemagglutinin-neuraminidase gene among virulent Newcastle disease viruses isolated in Iran. Iran J Vet Res 20:1–8. https://doi.org/10.22099/ijvr.2019.5135
Boroomand Z, Jafari RA, Mayahi M (2016) Molecular characterization and phylogenetic study of the fusion genes of Newcastle disease virus from the recent outbreaks in Ahvaz, Iran. VirusDisease 27:102–105. https://doi.org/10.1007/s13337-015-0299-z
Hosseini H, Langeroudi AG, Torabi R (2014) Molecular characterization and phylogenetic study of newcastle disease viruses isolated in Iran, 2010–2012. Avian Dis 58:373–376. https://doi.org/10.1637/10743-120713-reg.1
Langeroudi AG, Hosseini H, Karimi V et al (2014) Phylogenetic study base on matrix gene of Iranian Newcastle disease virus isolates, 2011–2012. Comp Clin Path 23:77–81. https://doi.org/10.1007/s00580-012-1573-8
Goudarzi H, Borm SVAN, Bashashati M et al (2019) Characterization and full genome sequencing of a velogenic Newcastle disease virus (NDV) strain Ck/IR/Beh/2011 belonging to subgenotype VII(L). Acta Virol 63:217–222. https://doi.org/10.4149/av_2019_206
Sabouri F, Vasfi Marandi M, Bashashati M (2018) Characterization of a novel VIIl sub-genotype of Newcastle disease virus circulating in Iran. Avian Pathol 47:90–99. https://doi.org/10.1080/03079457.2017.1376735
Nei M, Kumar S (2000) Molecular Evolution and Phylogenetics. Oxford University Press, New York
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution (NY) 39:783–791. https://doi.org/10.2307/2408678
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035. https://doi.org/10.1073/pnas.0404206101
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Funding
The research was funded by Razi Vaccine and Serum Research Institute for the project numbered 13-18-1851-070-97020-971049.
Author information
Authors and Affiliations
Contributions
AM: designed the study, performed the experiments, carried out the computer analyses and wrote the manuscript. MS: conceived the project, provided samples, performed some experiments and revised the manuscript. MHFM: conceived and supervised the project and also provided the funding. AS: provided samples and supervised AA: assisted in the experiments and provided samples. MMA: performed all the inoculations and pathogenicity assays. SAP: provided resources. SHEL: reviewed and revised the manuscript. AGL: conceived the study and provided resources. EA: provided samples, performed experiments, carried out computer analyses. MA: performed the pathogenicity assays including ICPI. ME: performed the Bayesian analysis and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Ethical approval
The Institutional Animal Care and Use Committee (IACUC) of Razi Vaccine and Serum Research Institute approved all animal experiments (permit code: RVSRI.REC.99.002).
Informed Consent
The present study did not involve any human subject.
Research Involving Human Participants and/or Animals
The present study did not involve any human subject. Animal handling procedures were performed in line with the national animal welfare regulations. The Institutional Animal Care and Use Committee (IACUC) of Razi Vaccine and Serum Research Institute approved all animal experiments (Permit Code: RVSRI.REC.99.002).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2021_2572_MOESM1_ESM.docx
Supplementary file 1 A list of NDV isolates sequenced in our laboratory as well as all the local (144 isolates) and several regional (70 isolates) sequences derived from GenBank, ViPR database and publications. The list includes all the Iranian submissions to date (as of September 2020), however, all the pigeon-derived sequences and most vaccinal strains were omitted. Only the longest GenBank submission was used if multiple submissions for one isolate had been made. This included submission for different genes of one isolate since the aim of study was identifying genotypes. The F sequences were used to make phylogenetic tree and conduct Bayesian analysis. Some sequences did not include the complete CDS, yet they were studied to find the closest genotypes. Genotypes for each of the isolates were carefully determined by phylogenetic anaysis in the current study. The list is primarily sorted by country and then arranged based on the year of collection. (DOCX 174 kb)
284_2021_2572_MOESM2_ESM.fas
Supplementary file 2 A FASTA file containing all the Iranian and regional NDV F gene sequences (except those isolated from Columbiformes) is available for download (341 isolates). The dataset will be regularly updated at our online repository. See text for URL. The sequences were added to the latest pilot NDV dataset of the international NDV consortium. The other datasets of the study can be obtained by unselecting the sequences that are not required for a particular analysis. For example, the first dataset is made of the consortium pilot file and the isolates mentioned in Table 1. All the short sequences that may have contributed to error during genetic and mino acid distance analysis were removed from the file. (FAS 573 kb)
284_2021_2572_MOESM3_ESM.pdf
Supplementary file 3 A philogenetic tree made using the first dataset of this study. The dataset itself was made by adding the F gene sequence of the 19 isolates of this study (marked with ●) to the pilot file of the international NDV consortium (125+19=144 sequences). All the isolates belonged to the subgenotype VII.1.1. The evolutionary history was inferred by using the Maximum Likelihood method and General Time Reversible model [46]. The bootstrap consensus tree was inferred from 1000 replicates [47]. A discrete Gamma distribution was used to model evolutionary rate differences among sites (4 categories (+G, parameter = 0.7347)). There were a total of 1662 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [24]. (PDF 133 kb)
284_2021_2572_MOESM4_ESM.tif
Supplementary file 4 A tree made using the second dataset of the current study (Supplementary File 2). The dataset was made by adding all the F gene sequences of the isolates of Supplementary File 1 to the pilot file of the international NDV consortium (total of 341 isolates). The Iranian NDV genotype VII.1.1 (formerly VIIl) formed a distinct cluster that has been consistent over a decade. Two Iraqi NDV isolated in 2012 also clustered within the Iranian group. Some Saudi Arabian VII.1.1 isolates were closely related to the Iranian VII.1.1 isolates, yet they formed a different cluster of their own (see Supplementary File 5 for genetic distances). The only VIId isolated from Iran was also shown in a different branch next to the VIId isolate included in the pilot dataset. The tree also shows the location of Iranian and regional subgenotype VII.2 NDV compared with the VII.2 isolates included in the pilot dataset of the international NDV consortium. The Iranian VII.2 isolates sided with the newly reported Belgian, Baghdadi and Israeli isolates closer than those from other countries (see Supplementary File 5 for distances). Some Iraqi VII.2 isolates from 2012 formed a different cluster. The evolutionary history was inferred using the Maximum Likelihood method and General Time Reversible model [46]. The bootstrap consensus tree was inferred from 1000 replicates [47]. A discrete Gamma distribution was used to model evolutionary rate differences among sites (4 categories (+G, parameter = 0.4727)). There were a total of 1662 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [24]. All isolates used in the current study are marked: ● Iran, ■ Iraq, ▼ Turkey, ▲ Saudi Arabia, ∆ UAE, ∇ Israel, □ Belgium, ♦ Pakistan, ◊ Indonesia. (TIF 2720 kb)
284_2021_2572_MOESM5_ESM.xlsx
Supplementary file 5 Estimates of evolutionary divergence between Iranian, regional and international NDV F gene sequences. Analyses were conducted using the Maximum Composite Likelihood model [48](500 bootstrap replications). The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The analysis involved 340 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 1662 positions in the final dataset. Evolutionary analyses were conducted in MEGA 7 [49]. (XLSX 691 kb)
Rights and permissions
About this article
Cite this article
Molouki, A., Sotani, M., Fallah Mehrabadi, M.H. et al. Predominance of Fourth Panzootic Newcastle Disease Virus Subgenotype VII.1.1 in Iran and Its Relation to the Genotypes Circulating in the Region. Curr Microbiol 78, 3068–3078 (2021). https://doi.org/10.1007/s00284-021-02572-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02572-z