Abstract
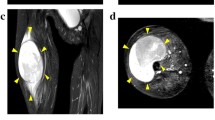
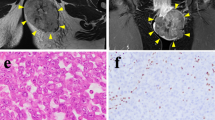
Spindle cell/sclerosing rhabdomyosarcoma (ssRMS) is a rare subtype of rhabdomyosarcoma (RMS) that has fascicular spindle cell and/or sclerosing morphology. SsRMS has a diverse molecular background and is categorized into three groups: congenital/infantile ssRMS with a gene fusion involving the NCOA2 and VGLL2, ssRMS with the MYOD1 mutation, and ssRMS with no recurrent identifiable genetic alterations. Because ssRMS is a newly defined disease concept of RMS, the optimal treatment methods have not been determined. This results in unfavorable prognosis and consequently signals the urgent need for continuous research. Patient-derived cell lines are essential tools in basic and translational research. However, only two ssRMS cell lines with the MYOD1 mutation have been reported to date. Thus, we established a novel ssRMS cell line named NCC-ssRMS2-C1 using a surgically resected tumor tissue from an adult ssRMS patient. NCC-ssRMS2-C1 cells retained the copy number alterations corresponding to the original tumor and are categorized into the group with no recurrent identifiable genetic alterations. NCC-ssRMS2-C1 cells demonstrated constant proliferation, spheroid formation, and capability for invasion in vitro, reflecting the malignant features of the original tumor tissue. In a drug screening test, ssRMS demonstrated remarkable sensitivity to romidepsin, trabectedin, actinomycin D, and bortezomib. Hence, we conclude that the NCC-ssRMS2-C1 cell line is the first ssRMS cell line which belongs to the group with no recurrent identifiable genetic alterations, and it will be a useful resource in both basic and translational studies for ssRMS.





Similar content being viewed by others
References
WHO Classification of Tumours of Soft Tissue and Bone. 5th ed. Lyon: IARC; 2020.
Dziuba I, Kurzawa P, Dopierala M, Larque AB, Januszkiewicz-Lewandowska D. Rhabdomyosarcoma in children - current pathologic and molecular classification. Pol J Pathol. 2018;69:20–32.
Pappo AS, Dirksen U. Rhabdomyosarcoma, ewing sarcoma, and other round cell sarcomas. J Clin Oncol. 2018;36:168–79.
Rekhi B, Singhvi T. Histopathological, immunohistochemical and molecular cytogenetic analysis of 21 spindle cell/sclerosing rhabdomyosarcomas. APMIS. 2014;122:1144–52.
Alaggio R, Zhang L, Sung YS, et al. A molecular study of pediatric spindle and sclerosing rhabdomyosarcoma: identification of novel and recurrent VGLL2-related fusions in infantile cases. Am J Surg Pathol. 2016;40:224–35.
Mosquera JM, Sboner A, Zhang L, et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–50.
Rekhi B, Upadhyay P, Ramteke MP, Dutt A. MYOD1 (L122R) mutations are associated with spindle cell and sclerosing rhabdomyosarcomas with aggressive clinical outcomes. Mod Pathol. 2016;29:1532–40.
Szuhai K, de Jong D, Leung WY, Fletcher CD, Hogendoorn PC. Transactivating mutation of the MYOD1 gene is a frequent event in adult spindle cell rhabdomyosarcoma. J Pathol. 2014;232:300–7.
Zhao Z, Yin Y, Zhang J, et al. Spindle cell/sclerosing rhabdomyosarcoma: case series from a single institution emphasizing morphology, immunohistochemistry and follow-up. Int J Clin Exp Pathol. 2015;8:13814–20.
Yasui N, Yoshida A, Kawamoto H, Yonemori K, Hosono A, Kawai A. Clinicopathologic analysis of spindle cell/sclerosing rhabdomyosarcoma. Pediatr Blood Cancer. 2015;62:1011–6.
Tseng YY, Boehm JS. From cell lines to living biosensors: new opportunities to prioritize cancer dependencies using ex vivo tumor cultures. Curr Opin Genet Dev. 2019;54:33–40.
Ben-David U, Siranosian B, Ha G, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–30.
Ben-David U, Beroukhim R, Golub TR. Genomic evolution of cancer models: perils and opportunities. Nat Rev Cancer. 2019;19:97–109.
Saito S, Morita K, Kohara A, et al. Use of BAC array CGH for evaluation of chromosomal stability of clinically used human mesenchymal stem cells and of cancer cell lines. Hum Cell. 2011;24:2–8.
Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23.
Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–61.
Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–61.
Goodspeed A, Heiser LM, Gray JW, Costello JC. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res. 2016;14:3–13.
Yoshimatsu Y, Noguchi R, Tsuchiya R, et al. Establishment and characterization of NCC-ssRMS1-C1: a novel patient-derived spindle-cell/sclerosing rhabdomyosarcoma cell line. Hum Cell. 2020;33:886–93.
Schleicher S, Grote S, Malenke E, et al. Establishment and characterization of a sclerosing spindle cell rhabdomyosarcoma cell line with a complex genomic profile. Cells. 2020;9:2668.
Bairoch A. The cellosaurus, a cell-line knowledge resource. J Biomol Tech. 2018;29:25–38.
Sin Y, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of a novel alveolar rhabdomyosarcoma cell line, NCC-aRMS1-C1. Hum Cell. 2020;33:1311–20.
Capes-Davis A, Reid YA, Kline MC, et al. Match criteria for human cell line authentication: where do we draw the line? Int J Cancer. 2013;132:2510–9.
Tsuchiya R, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of NCC-DDLPS3-C1: a novel patient-derived cell line of dedifferentiated liposarcoma. Hum Cell. 2021;34:1008–18.
Billiau A, Edy VG, Heremans H, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–5.
Walther C, Mayrhofer M, Nilsson J, et al. Genetic heterogeneity in rhabdomyosarcoma revealed by SNP array analysis. Genes Chromosomes Cancer. 2016;55:3–15.
Yang X, Yang S, Wang C, Kuang S. The hypoxia-inducible factors HIF1alpha and HIF2alpha are dispensable for embryonic muscle development but essential for postnatal muscle regeneration. J Biol Chem. 2017;292:5981–91.
Demetri GD, von Mehren M, Jones RL, et al. Efficacy and safety of trabectedin or dacarbazine for metastatic liposarcoma or leiomyosarcoma after failure of conventional chemotherapy: results of a phase III randomized multicenter clinical trial. J Clin Oncol. 2016;34:786–93.
Iyer SP, Foss FF. Romidepsin for the treatment of peripheral T-Cell lymphoma. Oncologist. 2015;20:1084–91.
Bharathy N, Berlow NE, Wang E, et al. Preclinical rationale for entinostat in embryonal rhabdomyosarcoma. Skelet Muscle. 2019;9:12.
Bharathy N, Berlow NE, Wang E, et al. The HDAC3-SMARCA4-miR-27a axis promotes expression of the PAX3:FOXO1 fusion oncogene in rhabdomyosarcoma. Sci Signal. 2018;11:eaau7632.
Tomoyasu C, Kikuchi K, Kaneda D, et al. OBP801, a novel histone deacetylase inhibitor, induces Mphase arrest and apoptosis in rhabdomyosarcoma cells. Oncol Rep. 2019;41:643–9.
Gandolfi S, Laubach JP, Hideshima T, Chauhan D, Anderson KC, Richardson PG. The proteasome and proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev. 2017;36:561–84.
Bersani F, Taulli R, Accornero P, et al. Bortezomib-mediated proteasome inhibition as a potential strategy for the treatment of rhabdomyosarcoma. Eur J Cancer. 2008;44:876–84.
Peron M, Bonvini P, Rosolen A. Effect of inhibition of the ubiquitin-proteasome system and Hsp90 on growth and survival of rhabdomyosarcoma cells in vitro. BMC Cancer. 2012;12:233.
Acknowledgements
We thank Drs. E. Kobayashi, S. Iwata, S. Fukushima, M. Nakagawa, T. Komatsubara, C. Sato (Department of Musculoskeletal Oncology), and Drs. N. Kojima, J. Kashima, M. Arakaki (Department of Diagnostic Pathology), National Cancer Center Hospital, for sampling tumor tissue specimens from surgically resected materials. We also appreciate the technical assistance provided by Ms. Y. Kuwata (Division of Rare Cancer Research). We appreciate the technical support provided by Ms. Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). We would also like to thank Editage (www.editage.jp) for their help with English language editing and their constructive comments on the manuscript. This research was technically assisted by the Fundamental Innovative Oncology Core in the National Cancer Center.
Funding
This research was supported by the Japan Agency for Medical Research and Development (Grant number 20ck0106537h0001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The ethical committee of the National Cancer Center approved the use of clinical materials for this study (Approval number 2004–050).
Informed consent
Written informed consent for publication was provided by the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13577_2021_569_MOESM3_ESM.tif
Supplementary file3 Supplementary Fig. 3 Growth curves for the IC50 value calculations of 24 anti-cancer agents (TIF 598 kb)
Rights and permissions
About this article
Cite this article
Tsuchiya, R., Yoshimatsu, Y., Noguchi, R. et al. Establishment and characterization of NCC-ssRMS2-C1: a novel patient-derived cell line of spindle cell/sclerosing rhabdomyosarcoma. Human Cell 34, 1569–1578 (2021). https://doi.org/10.1007/s13577-021-00569-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00569-1