Abstract
Scientists have developed and deployed successive generations of genome engineering technologies for use in plants, including meganucleases, zinc finger nucleases, TAL effector nucleases, and CRISPR nucleases. Each of these tools has been hailed as potentially revolutionary, capable of providing more efficient and precise ways to modify plant genomes toward improving agronomic traits or making fundamental discoveries. The CRISPR nucleases, in particular, have accelerated the pace of innovation and expanded the boundaries of what is achievable within the plant research space. This review will take care to discuss current plant genome engineering technologies, covering both well-established and up-and-coming tools, as well as describe potential and real-world applications.
Similar content being viewed by others
Room for improvement: generations of genome engineering tools
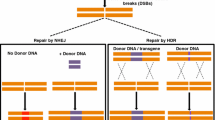
The process of introducing targeted modifications into a plant genome involves three common steps: recognition of a target DNA sequence, induction of a break, and repair. First, the sequence recognition module of the engineered nuclease recognizes the target DNA sequence. Next, the nuclease binds to the target DNA sequence and creates either a double-strand break (DSB) or single-strand break. Lastly, the DNA break is mended—either by the endogenous DNA repair pathways or by an engineered mechanism. The major DNA repair pathways include non-homologous end joining (NHEJ) and homology-directed repair (HDR) (Symington and Gautier 2011). A notable difference between these pathways is that while NHEJ is an error-prone repair process and often results in the introduction of mutations, such as small insertions and deletions (indels), HDR results in a precise repair. These basic principles underpin all of the genome-editing technologies currently in use, with the key differences between tools distilling down to additional design features, such as modularity, specificity, efficiency, ease of delivery, and the interplay between these factors. There is no ‘best’ genome engineering tool, just the best tool for a specific job. This is especially true as it pertains to more recently discovered tools, e.g., those under the ever-expanding CRISPR-Cas umbrella.
Meganucleases
Meganucleases, also referred to as homing endonucleases, were first described in the 1990s. In the organisms where they naturally occur, meganucleases are encoded by mobile genetic elements, either introns or inteins, and use double-strand breaks to propagate through the genome as part of selfish DNA elements (Voytas 2013; Smith et al. 2006; Paques and Duchateau 2007). Meganucleases are often small proteins that function as homodimer complexes, recognizing large stretches of DNA (20–40 nucleotides) through a site intrinsic to the protein, and subsequently creating a double-strand break (Voytas 2013; Puchta and Fauser 2014). Well-known meganucleases include the yeast mitochondria-derived protein I-SceI and the green algae chloroplast-derived protein I-CreI. Because existing meganuclease binding sites are rare in genomes of interest, meganucleases have been engineered to recognize new target sequences (Seligman et al. 2002; Sussman et al. 2004; Rosen et al. 2006). The relatively long recognition sequence results in higher specificity and lower off-target cutting. However, due to the difficulty of modulating meganucleases to accommodate new specificities, engineered meganucleases have had a more limited application compared to other sequence-specific nucleases (Daboussi et al. 2015).
Zinc finger nucleases
Zinc finger nucleases (ZFNs) are artificial sequence-specific nucleases that realized a breakthrough in programmable nucleases (Kim et al. 1996). ZFNs are engineered by fusing an array of zinc finger DNA-binding domains to the non-specific cleavage domain of the restriction endonuclease FokI (Chen et al. 2019). Within the zinc finger array, each domain recognizes a 3-nucleotide target sequence; thus, the makeup and order of the domains can be modified to match the desired site. In the early 2000s, ZFNs were used for sequence-specific mutagenesis in tobacco, which was the first time an engineered endonuclease recognized and cleaved chromosomal DNA ( Lloyd et al. 2005; Wright et al. 2005). Despite these early advances, the application of ZFNs remained limited in plants due to the challenging nature of design and construction, as well as the restricted availability of targeting sites in genomes when compared to more recently developed genome engineering tools.
TAL effector nucleases
Similar to ZFNs, TAL effector nucleases (TALENs) are artificially engineered proteins that are composed of a modular array of DNA-binding domains, fused to the non-specific cleavage domain of FokI. The TALEN DNA-binding domain array design is derived from transcription activator-like (TAL) effectors, a family of proteins which uses such arrays to carry out their function. TAL effectors are secreted by the plant-pathogenic bacteria Xanthomonas spp. into plant host cells during infection, where they bind DNA sequences upstream of target genes in a sequence-specific manner, modulating expression of these genes to enhance infection (Mak et al. 2013). The DNA binding specificity of TAL effectors is encoded within tandem repeated amino acid sequences, with one repeat binding to a single DNA base (Mak et al. 2013). In TALENs, these repeats are arrayed to target specific sequences of interest, leveraging the natural modularity—and elegance—of TAL effectors to achieve genome engineering. Gene targeting reagents using TAL effector scaffolds have included not only TALENs, but also gene-specific activators and repressors (Geissler et al. 2011; Zhang et al. 2011; Mahfouz et al. 2012). Compared to meganucleases and ZFNs, TALENs are more programmable and have been more widely used in plant genome engineering. However, the high number of repeats makes the construction of TALENs and their delivery into plant cells challenging.
CRISPR-Cas nucleases
Similar to TALENS, the CRISPR-Cas system also borrows its elegance from nature. CRISPR-Cas nucleases were first characterized as part of the bacterial and archaeal adaptive immune system. Here, clustered regularly interspaced short palindromic repeats (CRISPR) encode for ‘spacer’ RNA molecules that form complexes with CRISPR-associated (Cas) nucleases and direct these proteins to degrade foreign nucleic acids. In these natural systems, the spacer sequences are derived from fragments of bacteriophages that had previously infected the prokaryote lineage, with the targeting specificity of the system dependent on the simple rules of nucleotide base-pairing. In order for a target DNA site to be recognized and cleaved, the sequence also needs to include a distal or proximal short sequence–specific motif, termed the protospacer adjacent motif (PAM); this requirement ensures that the prokaryote employing the CRISPR-Cas system will not target its own genome, as these motifs are not present between endogenous spacer sequences.
The CRISPR-Cas9 system from Streptococcus pyogenes (CRISPR-SpCas9) was the first to be developed for genome engineering (Barrangou et al. 2007; Bhaya et al. 2011; Terns and Terns 2011; Sorek et al. 2013; Koonin et al. 2017; Shmakov et al. 2017), and ‘CRISPR-Cas9’ often refers to this particular tool. To avoid confusion, herein, CRISPR-Cas9 is only used to refer to features common to CRISPR-SpCas9 and its orthologs; in other cases, the origin species is noted. The engineered CRISPR-Cas9 system has two components: the (1) Cas9 nuclease and (2) a single guide RNA (sgRNA), consisting of a fusion of two RNA molecules—the spacer-containing CRISPR RNA (crRNA) and the transactivating crRNA (tracrRNA), which is required for maturation of the former moiety. The sgRNA directs the nuclease complex to a specific target DNA site, initiating the cleavage of the complementary DNA sequence (Chen et al. 2019). Cas9 possesses a bi-lobed architecture, with the smaller nuclease lobe containing two nuclease domains, RuvC and HNH, which cleave DNA strands that are complementary and non-complementary, respectively (Gasiunas et al. 2012; Jinek et al. 2012). In 2013, three independent groups established the CRISPR-SpCas9 system for use in rice, wheat, tobacco (Nicotiana benthamiana), and Arabidopsis (Li et al. 2013; Nekrasov et al. 2013; Shan et al. 2013), and the development and application of this system has since progressed rapidly (Chen et al. 2019; Zhang et al. 2019; Wada et al. 2020). The CRISPR-Cas9 system has many advantages over other plant genome engineering tools (Andolfo et al. 2016)—including relative ease of design, cloning, and delivery into plant cells—which has led to the wide adoption of this technology and the development of myriad related technologies.
There are multiple strategies to deliver CRISPR-Cas reagents into plant cells, including stable expression, transient expression, and DNA-free delivery. Genome engineering using stable expression of CRISPR-Cas DNA has been widely and successfully applied, through the tried-and-true methods of plant transformation and selection. However, some applications are incompatible with CRISPR constructs or marker genes integrated into the genome. In these cases, transgene-free derivatives can be obtained from stable transgenic plants through segregation over successive generations of plant propagation, or alternative methods for CRISPR-Cas reagent delivery can be used. In the transient CRISPR-Cas DNA delivery method, the normal selection steps are eliminated such that some regenerated plants are edited without any DNA integration, although this strategy can complicate downstream plant screening (Andersson et al. 2017; Lin et al. 2018; Chen et al. 2018). Additional methods involve delivering transcripts encoding CRISPR-Cas reagents directly into embryos to generate edited plants, albeit with lower editing efficiency (Zhang et al. 2011). Recently, an RNA virus–based vector system has been engineered for transgene-free delivery of the CRISPR-Cas cassette for efficient genome engineering in plants (Ma et al. 2020). Additionally, a DNA-free system has been developed in plants using Cas9-sgRNA ribonucleoprotein (RNP) complexes, which have been shown to be as efficient as stable delivery, as well as to exhibit a low off-target frequency (Woo et al. 2015; Svitashev et al. 2016; Liang et al. 2017, 2018).
Nothing was the same: the CRISPR-Cas revolution
Cas9 is not the only CRISPR-associated protein—it is just one family of proteins among many other CRISPR protein families. There are two major classes of CRISPR systems, differentiated by the method of CRISPR RNA processing: if the pre-CRISPR RNA processing and interference stages are accomplished by one single multifunctional protein, the CRISPR system is categorized as a class 2 system; otherwise, it is categorized as a class 1 system (Makarova et al. 2015). Each of these classes is divided into multiple types according to their signature proteins: type I, III, and IV belong to class 1, with Cas3, Cas10, and Csf1 as their respective signature proteins, while type II (Cas9), type V (Cas12a–e, Cas12g–i, and Cas14a–c), and type VI (Cas13a–d) belong to class 2 (Makarova et al. 2015). Recent research has expanded access to CRISPR-Cas systems beyond Cas9, elevating new systems with distinct advantages and disadvantages.
SpCas9 variants
The SpCas9 protein has been extensively engineered to broaden the PAM compatibility, enhance specificity, and confer new functionality (Karvelis et al. 2017). SpCas9 requires a 5′-NGG-3′ PAM, where ‘N’ is any nucleotide, limiting which genome positions can be targeted, especially in AT-rich species. So far, rational engineering of SpCas9 has resulted in the generation of new Cas9 variants with four alternate PAM preferences (Kleinstiver et al. 2015; Nishimasu et al. 2018). One of these engineered Cas9 proteins has already been used to edit rice and Arabidopsis plants (Endo et al. 2019; Ge et al. 2019; Hua et al. 2019; Niu et al. 2020; Zhong et al. 2019), pointing to the promise of this strategy to expand the availability of target sites. In addition, there are a number of SpCas9 proteins that have been rationally engineered to have enhanced specificity for their target sequence, thus reducing off-target activity (Zhang et al. 2017a, 2017b; Abudayyeh et al. 2016; Chen et al. 2017). Similar variants have been developed using directed evolution (Casini et al. 2018; Hu et al. 2018; Lee et al. 2018). Other engineered SpCas9 variants include nuclease domain mutants, which result in new functional characteristics. Inactivation of a single nuclease domain results in a nuclease that only causes a single-strand break, termed a ‘nickase’ (nCas9), whereas inactivation of both domains produces a deactivated Cas9 protein (dCas9) (Jiang and Doudna 2017).
SpCas9 orthologs
Complementary to SpCas9 engineering efforts, there has been a lot of research around discovering Cas9 orthologs from other species, as evolution has already worked to produce variants with diverse functionalities. So far, Cas9 orthologs from a number of bacterial species have been characterized, including NmCas9 from Neisseria meningitidis (Hou et al. 2013), SaCas9 from Staphylococcus aureus (Ran et al. 2015), StCas9 from Streptococcus thermophilus (Müller et al. 2016), FnCas9 from Francisella novicida (Hirano et al. 2016), and CjCas9 from Campylobacter jejuni (Kim et al. 2017). These Cas9 orthologs have different PAM preferences, expanding the range of sequences that can be targeted (Wada et al. 2020). In addition, the genes encoding most of these proteins are smaller than SpCas9, providing an advantage for delivery into plant cells (Wada et al. 2020). Some of the Cas9 orthologs, such as NmCas9, bind longer target sequences, conferring additional specificity (Hou et al. 2013). Having a collection of Cas9 orthologs to choose from is also beneficial when designing orthogonal gene targeting experiments, as these nucleases can be used simultaneously to target different sites in the genome ( Steinert et al. 2015 Puchta 2017).
Cas12a (Cpf1)
Cas12a—formerly known as Cpf1, CRISPR from Prevotella and Francisella 1—is a class 2 type V nuclease that has broad utility in genome engineering. Cas12a functions in a similar fashion to Cas9, forming a complex with a sgRNA to target specific DNA sequences to create double-stranded breaks. Cas12a requires a T-rich PAM sequence, as compared to the G-rich PAM of Cas9, broadening the species and sites that can be targeted. The sgRNA architecture that is required for Cas12a is shorter than the Cas9 sgRNA architecture, making the Cas12a sgRNAs easier to synthesize, multiplex, and engineer. Moreover, Cas12a possesses RNase activity, meaning it can self-process a polycistronic CRISPR sgRNA array for multiplexed genome editing (Ran et al. 2015; Yamano et al. 2016). Cas12a also cuts DNA in a staggered fashion, creating an overhang which may promote repair and thus hypothesized to improve HDR efficiencies (Zetsche et al. 2015).
There are a number of Cas12a orthologs currently in use, including proteins from Francisella novicida (FnCas12a), Acidaminococcus sp. (AsCas12a), and Lachnospiraceae bacterium (LbCas12a) (Zetsche et al. 2015). Some of these Cas12a orthologs have been found to be more specific than SpCas9 in several biological systems (Kim et al. 2016; Kleinstiver et al. 2016b; Tang et al. 2017; Zhong et al. 2018). Characterization of diverse Cas12a orthologs has identified nucleases with more varied PAMs beyond the standard site (Marshall et al. 2018). Similar to Cas9, Cas12a variants have been engineered to recognize different PAMs, with more than five distinct PAM sites currently available (Gao et al. 2017; Li et al. 2018; Zhong et al. 2018). Moreover, multiple versions of catalytically inactive Cas12a (dCas12a) have been engineered and repurposed for different applications (Zetsche et al. 2015; Yamano et al. 2016; Tang et al. 2017; Zhang et al. 2017b). So far, LbCas12a has been most widely deployed, having been used to successfully create edits in rice (Begemann et al. 2017a, 2017b; Xu et al. 2017; Yin et al. 2017). Continual advancements in the application of Cas12a for genome engineering will position this nuclease as complimentary to Cas9.
Cms1
In addition to Cas12a, another group of class 2 type V enzymes, termed Cms1s—CRISPR from Microgenomates and Smithella 1—efficiently generate indel mutations in rice (Begemann et al. 2017a, 2017b). Cms1 nucleases are smaller than both Cas9 and Cas12a nucleases and do not require a transactivating crRNA (Begemann et al. 2017a, 2017b). Cms1 nucleases also have an AT-rich PAM site requirement, which can offer obvious situational advantages as compared to other CRISPR-Cas nucleases (Begemann et al. 2017a, 2017b). Although Cms1 nucleases are still being developed for broader application, the differences in the function of these enzymes make them a potentially invaluable addition to the plant genome engineering toolbox.
Cas13a
Cas13a, formerly known as C2c2, has RNase activity and targets single-stranded RNA for degradation (Abudayyeh et al. 2016). Instead of a PAM, Cas13a requires a protospacer flanking site to induce a single-strand break. Interestingly, Cas13a also shows non-specific RNase activity that cleaves collateral RNA following initial binding to its target RNA in vitro and in bacteria (Abudayyeh et al. 2016; Gootenberg et al. 2017). This particular feature has led to the development of SHERLOCK, the specific high-sensitivity enzymatic reporting unlocking method, which can be used to detect specific RNA and DNA sequences (Abudayyeh et al. 2016). The CRISPR-Cas13a system has been used to decrease the expression of endogenous genes in rice and tobacco (N. benthamiana) (Abudayyeh et al. 2016; Aman et al. 2018). This system has also been engineered to interfere with RNA-guided immunity in Arabidopsis and tobacco (N. benthamiana) (Aman et al. 2018). These specific applications go beyond the possibilities presented by CRISPR-Cas9. Testing and developing more Cas13a orthologs and variants in plants will add additional RNA-targeting tools for transcriptional regulation, RNA base editing, RNA tracking, functional studies, pathogen detection, and disease control (Zhang et al. 2019).
Base editors
Base editing is a type of genome engineering that involves making specific nucleotide substitutions without reliance on the formation of a double-strand break or a donor template. To date, most base editing systems have involved linking base editing enzymes to nCas9, which is able to guide the enzyme to target sites where activity is then proximity-induced. The cytosine base editor (CBE) systems mediate the conversion of C to T in genomic DNA, with the base-editing enzymes including the combination of a cytidine deaminase and a uracil glycosylase inhibitor (Komor et al. 2016). Different CBE systems have leveraged different cytidine deaminase enzymes, with the rat APOBEC1 enzyme the most widely used in plants (Hess et al. 2017). C-to-G base editor systems have also been applied in plants, relying on nCas9 linked to CBE variants and uracil DNA N-glycosylase. The adenine base editor (ABE) systems mediate the conversion of A to G in genomic DNA (Gaudelli et al. 2017). This system links an nCas9 to an adenosine deaminase, with the Escherichia coli TadA enzyme the most commonly used in plants (Gaudelli et al. 2017). Base-editing systems offer several advantages over non-DSB-mediated genome editing in plants, including greater efficiency and precision. Moreover, multiplexed base editing is less likely to result in chromosomal rearrangements—which can happen with standard multiplexed editing. Additional research with a focus on the improvement of the efficiency and specificity of these technologies, as well as their compatibility with multiplexing, will underpin more successful precision breeding efforts.
Prime editors
Prime editing is one of the latest additions to the CRISPR-Cas toolbox. This system performs RNA template-based DNA modifications using an engineered reverse transcriptase and is able to install small additions, deletions, and all 12 possible nucleotide conversions (Anzalone et al. 2019). A prime editor involves a fusion of nCas9 and reverse transcriptase, programmed with prime editing guide RNAs (pegRNAs) that encode the desired edit (Q. Lin et al. 2020). Prime editing was first applied in rice and wheat, with prime-edited rice plants obtained at high frequencies (Lin et al. 2020). Several subsequent studies have reported varying efficiencies in rice and other species (Butt et al. 2020; Hua et al. 2020; Jiang et al. 2020; Veillet et al. 2020). A vector system for prime editing was recently developed and validated for use in tobacco (N. benthamiana), rice, and Arabidopsis (Wang et al. 2021). The programmable and precise nature of prime editing makes it a powerful tool for plant genome engineering, and there will undoubtedly be an explosion of studies that apply and improve upon this tool.
Views: applications of genome editing in plants
The profusion of available genome engineering tools, as well as improvements in editing efficiency and precision, has led to an expansion of potential applications for these technologies. More recently developed tools have been used to induce homology-directed repair and edit multiple sites in parallel—approaches that stand to radically alter the feasibility of using genome engineering for basic research and crop improvement. In addition to discussing these advancements, this section provides a conceptual introduction to the types of edits that one could conceive of, using any number of the current technologies (Fig. 1).
Types of edits that can be generated with genome engineering technologies. Range of edits that can be made using targeted insertions or deletions (indels), base edits, prime edits, or homology-directed repair (HDR). Edits can be generated in coding or genic regions (top), including gene knockouts, modifications, in-frame fusions, and replacements. Edits can also be targeted to non-coding regions (middle); this includes cis element modifications and insertions, as well as modifications to upstream open reading frames (uORF) or whole promoter replacement. Edits can also span coding and non-coding regions (bottom), resulting in whole-gene deletion or even larger chromosome segment deletions.
Homology-directed repair
The homology-directed repair (HDR) pathway uses a template with homology to a double-strand break site to patch DNA damage. The template can be derived from the genome, such as from a sister chromatid, or from exogenously supplied DNA. For the type of HDR used in genome engineering, a repair template with desired sequence modifications is provided along with editing machinery, and the endogenous HDR pathway incorporates these changes into the break site (Salsman and Dellaire 2017). HDR can be used to introduce specific point mutations, as well as to insert, or replace, desired sequences at a target DNA site. Precise gene modifications—such as knock-ins and replacements—facilitate breeding by introducing new alleles without linkage drag or generating allelic variants that do not exist naturally. However, it is still quite challenging to perform HDR-mediated gene targeting in plants due to the low efficiency of HDR and the limitations of donor template delivery in plant cells. One approach to increase HDR efficiency is to increase the amount of donor DNA delivered to the cell. Toward this end, a geminivirus-based DNA replicon has been used to increase the number of repair templates and improve efficiency across multiple plant species (Čermák et al. 2015; Butler et al. 2016; Wang et al. 2017; Dahan-Meir et al. 2018; Hummel et al. 2018). HDR efficiency can also be improved by changing the design or delivery method of the donor template. These modifications include increasing the length of homology arms or including a tag for targeting (Carlson-Stevermer et al. 2017; Ma et al. 2017; Aird et al. 2018; Ghanta et al. 2021).
Multiplex editing
Plant traits are rarely determined by a single gene but are dependent on the contributions of multiple genes. Thus, the goals of genome engineering go beyond editing single sites to editing multiple sites simultaneously, called multiplex editing. Multiplex editing has a number of exciting applications for genome engineering: it can be used to create multi-gene knockouts, chromosomal deletions and translocations, gene knock-ins, and quantitative variation across multiple sites (Salsman and Dellaire 2017). Of all of the available gene editing tools, the CRISPR-Cas system is by far the most amenable to multiplex editing, as multiple sgRNAs can be expressed along with a single nuclease to achieve editing at many distinct target sites (Hashimoto et al. 2018): Zsögön et al. 2018; Najera et al. 2019.) There are many methods to multiplex sgRNA expression, including using multiple tandem expression cassettes, or expressing sgRNAs as a polycistronic transcript under the control of a single promoter. In the latter case, the sgRNAs can be interspersed with ribozyme sites (Gao and Zhao 2014), Csy4 recognition sites (Čermák et al. 2017), or transfer RNA sequences (Xie et al. 2015)—all of which allow for processing in the plant cell to release mature sgRNAs for editing (Chen et al. 2019). In plants, CRISPR-Cas multiplex editing has been achieved in multiple species, with a recent publication reporting one-shot generation of 8× N. benthamiana and 12× Arabidopsis mutants (Stuttmann et al. 2021).
Protein-coding edits
Genome engineering tools have been primarily used to make edits in coding regions. To create a gene knockout, an edit can be targeted to an exon upstream of the protein active site residues. In this case, small insertions, deletions, or base changes can cause a frameshift mutation, leading to an early stop codon and a non-functional protein (Fig. 1). In cases where the protein structure is flexible enough to allow for modification, targeted deletions or precise modifications within genes can also be used to alter protein activation or repression sites (Fig. 1). Targeted insertions via HDR can be used to generate in-frame fusion proteins at the endogenous locus, a creative strategy to study the biological function of a given protein (Wang et al. 2017) (Fig. 1). Similarly, HDR-mediated insertions can be used to make gene replacements, including allele swaps (Fig. 1). Although not strictly protein-coding edits, tandem edits can be used to make mutations that span coding and non-coding regions, leading to whole-gene deletions or chromosomal segment deletions (Fig. 1) (Zhou et al. 2014; Belhaj et al. 2015). In the latter case, larger deletions can target clusters of genes with related functions, an especially powerful approach where there is predicted functional redundancy. All of these types of edits can be used to create desired phenotypes or study the function of a gene of interest.
Non-coding edits
Genome-editing technologies can also be used to create modifications in cis-regulatory regions to alter gene regulation. This strategy has primarily focused on promoter sequences, such as replacing whole promoters via HDR or editing specific cis-regulatory elements (Piatek et al. 2015; Peng et al. 2017) (Fig. 1). In tomato, researchers edited the promoter regions of quantitative trait–related genes, creating a continuum of variation and leading to the selection of artificial alleles with improved traits (Rodríguez-Leal et al. 2017). Similar work on other agronomic genes of interest will allow for an exploration of dosage effects, leading to a more precise understanding of the relationship between gene expression and phenotypic variation. Cis-regulatory editing will prove instrumental for engineering plants with desirable characteristics, especially in cases where specific gene overexpression is the most promising route and natural variation is limiting. In these cases, it will be possible to insert enhancer sequences, or other cis-elements that lead to promoter upregulation, in order to increase transcription of a target gene (Fig. 1). In addition to promoter engineering, gene regulation can also occur at the translation level, such as upstream open reading frames (uORFs) (Fig. 1). uORFs are well-characterized cis-elements, widespread among plant mRNAs, which often negatively regulate translation and mRNA decay (von Arnim et al. 2014). It has been shown that CRISPR-Cas targeting of a uORF in lettuce was able to significantly alter the metabolic profile of the leaf (Zhang et al. 2018). Additional research targeting uORFs is a promising strategy to alter phenotypes of interest and create plant varieties with desirable characteristics without making modifications to protein-coding regions.
Off-target edits
In addition to intended mutations, off-target mutations can also occur with the application of genome-editing tools. With all gene editing tools, there are known trade-offs between editing specificity and other priorities, such as ease of reagent design, cloning, delivery, and efficiency. Minimizing the likelihood of off-target mutations is one priority that must be balanced against other priorities and weighed accordingly based on research aims. Whereas off-target edits may not be such a concern when engineering plants to study the function of a particular protein in the lab, off-target edits are of critical importance when producing varieties that you may want to bring to market. Although off-target edits have often been discussed as a non-concern when applying CRISPR-Cas systems, studies that have performed whole-genome sequencing to detect off-target mutations resulting from the application of Cas9 or Cas12a nucleases have revealed that both of these nucleases have low incidence of off-targeting (Feng et al. 2014; Tang et al. 2018; Li et al. 2019).
Based on large-scale editing or binding experiments undertaken in non-plant systems, CRISPR-Cas off-target edits can be largely mitigated by prioritizing target specificity during sgRNA design by ensuring that your target sequence is unique and does not occur elsewhere in the genome, either as an exact match or with a low level of mismatches (Kim et al. 2016; Luo et al. 2019; Specht et al. 2020). Although this may be straightforward in species with small genomes, this may prove more challenging in species that have undergone whole-genome duplications. Moreover, specifically targeting edits to coding regions may be easier to accomplish than to non-coding regions, such as promoters, which can be highly repetitive.
Conclusions
With the benefit of hindsight, the pre-CRISPR-Cas9 part of the story of plant genome engineering can almost read like a prelude. However, it is worth considering that the principles that underpin the use of CRISPR-Cas9 to engineer plants—delivery of reagents into the cell, the breadth and diversity of desired edits, responsible use of genome engineering, etc.—have been under development for decades, with invaluable input from other fields of study. That said, it is difficult to overestimate the impact that CRISPR-Cas9 has had, and will continue to have, on plant science, not least of which is the development and application of related CRISPR-Cas technologies. Cas12a, Cms1s, and Cas13a are all essential components of the growing genome-editing toolbox, with distinct use cases that transcend the possibilities of Cas9 techniques alone. The imagined and real-world applications for all of these tools, and the available tools themselves, seem to multiply every day, rendering review articles such as this one out-of-date by the time of publication. The pace and impact of these developments make this an exceedingly exciting time to be working in the plant genome engineering space.
References
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353(6299):aaf5573. https://doi.org/10.1126/science.aaf5573
Aird EJ, Lovendahl KN, St Martin A, Harris RS, Gordon WR (2018) Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Comm Biol 1:54. https://doi.org/10.1038/s42003-018-0054-2
Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding S, Mahfouz M (2018) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19:1. https://doi.org/10.1186/s13059-017-1381-1
Andersson M, Turesson H, Nicolia A, Fält AS, Samuelsson M, Hofvander P (2017) Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep 36:117–128
Andolfo G, Iovieno P, Frusciante L, Ercolano MR (2016) Genome-editing technologies for enhancing plant disease resistance. Front Plant Sci 7:1813. https://doi.org/10.3389/fpls.2016.01813
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liuet DR (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576:149–157
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Begemann MB, Gray BN, January E, Gordon GC, He Y, Liu H, Wu X, Brutnell TP, Mockler TC, Oufattole M (2017a) Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci Rep 7:11606. https://doi.org/10.1038/s41598-017-11760-6
Begemann MB, Gray BN, January E, Singer A, Kesler DC, He Y, Liu H, Guo H, Jordan A, Brutnell TP, Mockler TC, Oufattole M (2017b) Characterization and validation of a novel group of type V, class 2 nucleases for in vivo genome editing. bioRxiv 2017. https://doi.org/10.1101/192799
Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V (2015) Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol 32:76–84
Bhaya D, Davison M, Barrangou R (2011) CRISPR-Cas systems in Bacteria and Archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273–297
Butler NM, Baltes NJ, Voytas DF, Douches DS (2016) Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front Plant Sci 7:1045. https://doi.org/10.3389/fpls.2016.01045
Butt H, Rao GS, Sedeek K, Aman R, Kamel R, Mahfouz M (2020) Engineering herbicide resistance via prime editing in rice. Plant Biotechnol J 18:2370–2372
Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K (2017) Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nature Comm 8:1711. https://doi.org/10.1038/s41467-017-01875-9
Casini A, Olivieri M, Petris G, Montagna C, Reginato G, Maule G, Lorenzin F, Prandi D, Romanel A, Demichelis F, Inga A, Ceresto A (2018) A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol 36:265–271
Čermák T, Baltes NJ, Čegan R, Zhang Y, Voytas DF (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16:232. https://doi.org/10.1186/s13059-015-0796-9
Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, Voytas DF (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29:1196–1217
Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA (2017) Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 550:407–410
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:667–697
Chen L, Li W, Katin-Grazzini L, Ding J, Gu X, Li Y, Gu T, Li Y, Gu T, Wang R, Lin X, Deng Z, McAvoy RJ, Gmitter FG Jr, Deng Z, Zhao Y, Let Y (2018) A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hort Res 5:13. https://doi.org/10.1038/s41438-018-0023-4
Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Czosnek H, Aharoni A, Levy AA (2018) Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 95:5–16
Daboussi F, Stoddard TJ, Zhang F (2015) Engineering meganuclease for precise plant genome modification. In: Zhang F, Puchta H, Thomson J (eds) Advances in new technology for targeted modification of plant genomes. Springer, New York, NY, pp 21–38. https://doi.org/10.1007/978-1-4939-2556-8_2
Endo M, Mikami M, Endo A, Kaya H, Itoh T, Nishimasu H, Nurek O, Toki S (2019) Genome editing in plants by engineered CRISPR–Cas9 recognizing NG PAM. Nature Plants. 5:14–17
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA. 111:4632–4637
Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang Z (2017) Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol 35:789–792
Gao Y, Zhao Y (2014) Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing: self-processing of ribozyme-flanked RNAs into guide RNAs. J Integr Plant Biol 56:343–349
Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109:E2579–E2586
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551:464–471
Ge Z, Zheng L, Zhao Y, Jiang J, Zhang EJ, Liu T, Gu H, Qu LJ (2019) Engineered xCas9 and SpCas9-NG variants broaden PAM recognition sites to generate mutations in Arabidopsis plants. Plant Biotechnol J 17:1865–1867
Geissler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, Boch J (2011) Transcriptional activators of human genes with programmable DNA-specificity. PLoS One 6(5):e19509
Ghanta KS, Chen Z, Mir A, Dokshin GA, Krishnamurthy P, Yoon Y, Gallant J, Xu P, Zhang XO, Ozturk A, Shin M, Idrizi F, Liu P, Gneid H, Lawson N, Rivera-Pérez JA, Sontheimer EJ, Watts JK, Mello C (2021) 5′ modifications improve potency and efficacy of DNA donors for precision genome editing. bioRxiv. https://doi.org/10.1101/354480
Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356:438–442
Hashimoto R, Ueta R, Abe C, Osakabe Y, Osakabe K (2018) Efficient multiplex genome editing induces precise, and self-ligated type mutations in tomato plants. Front Plant Sci 9:916. https://doi.org/10.3389/fpls.2018.00916
Hess GT, Tycko J, Yao D, Bassik MC (2017) Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol Cell 68:26–43
Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, Nishimasu H, Nureki O (2016) Structure and engineering of Francisella novicida Cas9. Cell 164:950–961. https://doi.org/10.1016/j.cell.2016.01.039
Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA (2013) Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci 110:15644–15649
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liuet DR (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556:57–63
Hua K, Jiang Y, Tao X, Zhu JK (2020) Precision genome engineering in rice using prime editing system. Plant Biotechnol J 18:2167–2169
Hua K, Tao X, Han P, Wang R, Zhu JK (2019) Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol Plant 12:1003–1014
Hummel AW, Chauhan RD, Cermak T, Mutka AM, Vijayaraghavan A, Boyher A, Starker CG, Bart R, Voytas DF, Taylor NJ (2018) Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol J 16:1275–1282
Jiang F, Doudna JA (2017) CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529. https://doi.org/10.1146/annurev-biophys-062215-010822
Jiang YY, Chai YP, Lu MH, Han XL, Lin Q, Zhang Y, Zhang Q, Zhou Y, Wang X-C, Gao C, Chen Q-J (2020) Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol 21:257. https://doi.org/10.1186/s13059-020-02170-5
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Karvelis T, Gasiunas G, Siksnys V (2017) Harnessing the natural diversity and in vitro evolution of Cas9 to expand the genome editing toolbox. Curr Opin Microbiol 37:88–94
Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS (2016) Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol 34:863–868
Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim JS (2017) In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nature Comm 8:14500. https://doi.org/10.1038/ncomms14500
Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93:1156–1160
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh JRJ, Aryee MJ, Joung K (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481–485
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung K (2016a) high-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529:490–495
Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK (2016b) Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol 34:869–874
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424
Koonin EV, Makarova KS, Zhang F (2017) Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78
Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, Lee K, Jung I, Kim S, Kim J-S (2018) Directed evolution of CRISPR-Cas9 to increase its specificity. Nature Comm 9:3048. https://doi.org/10.1038/s41467-018-05477-x
Li J, Manghwar H, Sun L, Wang P, Wang G, Sheng H, Zhang J, Liu H, Qin L, Rui H, Li B, Lindsey K, Daniell H, Jin S, Zhang X (2019) Whole genome sequencing reveals rare off-target mutations and considerable inherent genetic or/and somaclonal variations in CRISPR/Cas9-edited cotton plants. Plant Biotechnol J 17:858–868
Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691
Li S, Zhang X, Wang W, Guo X, Wu Z, Du W, Zhao Y, Xia L (2018) Expanding the scope of CRISPR/Cpf1- mediated genome editing in rice. Mol Plant 11:995–998
Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, Gao C (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nature Comm 8:14261. https://doi.org/10.1038/ncomms14261
Liang Z, Chen K, Zhang Y, Liu J, Yin K, Qiu JL, Gao C (2018) Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat Prot 13:413–430. https://doi.org/10.1038/nprot.2017.145
Lin CS, Hsu CT, Yang LH, Lee LY, Fu JY, Cheng QW, Wu FH, H HCW, Zhang Y, Zhang R, Chang WJ, Yu CT, Wang W, Liao LJ, Gelvin SB, Shih MC (2018) Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J 16:1295–1310
Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone AV, Raguram A, Doman JL, Liu DR, Gao C (2020) Prime genome editing in rice and wheat. Nat Biotechnol 38:582–585
Lloyd A, Plaisier CL, Carroll D, Drews GN (2005) Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci 102:2232–2237
Luo J, Chen W, Xue L, Tang B (2019) Prediction of activity and specificity of CRISPR-Cpf1 using convolutional deep learning neural networks. BMC Bioinformatics 20:332. https://doi.org/10.1186/s12859-019-2939-6
Ma M, Zhuang F, Hu X, Wang B, Wen XZ, Ji JF, Xi JJ (2017) Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-avidin/biotin-donor DNA system. Cell Res 27:578–581
Ma X, Zhang X, Liu H, Zhenghe Li Z (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nature Plant 6:773–779
Mahfouz MM, Li L, Piatek M, Fang X, Mansour H, Bangarusamy DK, Zhu JK (2012) Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol Biol 78:311–321
Mak ANS, Bradley P, Bogdanove AJ, Stoddard BL (2013) TAL effectors: function, structure, engineering and applications. Curr Opin Struct Biol 23:93–99
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garret RA, van der Oost J, Backofen R, Koonin EV (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736
Marshall R, Maxwell CS, Collins SP, Jacobsen T, Luo ML, Begemann MB, Gray BN, January E, Singer A, He Y, Beisel CL, Noireaux V (2018) Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol Cell 69:146-157.e3. https://doi.org/10.1016/j.molcel.2017.12.007
Müller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C (2016) Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol Therapy 24:636–644
Najera AV, Twyman RM, Christou P, Zhu C (2019) Applications of multiplex genome editing in higher plants. Curr Opin Biotechnol 59:93–102
Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nurkeki O (2018) Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361:1259–1262
Niu Q, Wu S, Li Y, Yang X, Liu P, Xu Y, Lang Z (2020) Expanding the scope of CRISPR/Cas9-mediated genome editing in plants using an xCas9 and Cas9-NG hybrid. J Integr Plant Biol 62:398–402
Paques F, Duchateau P (2007) Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Therap 7:49–66. https://doi.org/10.2174/156652307779940216
Peng A, Chen S, Lei T, Xu L, He Y, Wu L, Yao L, Zou X (2017) Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J 15:1509–1519
Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM (2015) RNA-guided transcriptional regulation in plants via synthetic dCas9-based transcription factors. Plant Biotechnol J 13:578–589
Puchta H (2017) Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr Opin Plant Biol 36:1–8
Puchta H, Fauser F (2014) Synthetic nucleases for genome engineering in plants: prospects for a bright future. The Plant J 78:727–741
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191
Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171:470–80.e8
Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM (2006) Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nuc Acid Res 34:4791–4800
Salsman J, Dellaire G (2017) Precision genome editing in the CRISPR era. Biochem Cell Biol 95:187–201. https://doi.org/10.1139/bcb-2016-0137
Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL (2002) Mutations altering the cleavage specificity of a homing endonuclease. Nuc Acid Res 30:3870–3879
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu J-L, Gao C (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV (2017) Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol 15:169–182
Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Edondo P, Blanco FJ, Bravo J, Montoya G, Paques F, Duchateau P (2006) A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nuc Acid Res 34:e149
Sorek R, Lawrence CM, Wiedenheft B (2013) CRISPR-mediated adaptive immune systems in bacteria and Archaea. Annu Rev Biochem 82:237–266
Specht DA, Xu Y, Lambert G (2020) Massively parallel CRISPRi assays reveal concealed thermodynamic determinants of dCas12a binding. Proc Natl Acad Sci 117:11274–11282
Steinert J, Schiml S, Fauser F, Puchta H (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. The Plant J 84:1295–1305
Stuttmann J, Barthel K, Martin P, Ordon J, Erickson JL, Herr R, Ferik F, Kretschmer C, Berner T, Keilwagen J, Marillonnet S, Bonas U (2021) Highly efficient multiplex editing: one-shot generation of 8x Nicotiana benthamiana and 12× Arabidopsis mutants. Plant J 106:8–22. https://doi.org/10.1111/tpj.15197
Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R Jr, Stoddard BL, Seligman LM (2004) Isolation and characterization of new homing endonuclease specificities at individual target site positions. J Mol Biol 342:31–41
Svitashev S, Schwartz C, Lenderts B, Young JK, Cigan AM (2016) Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nature Comm 7:13274
Symington LS, Gautier J (2011) Double-Strand Break End Resection and Repair Pathway Choice. Annu Rev Genet 45:247–271
Tang X, Liu G, Zhou J, Ren Q, You Q, Tian L, Xin X, Zhong Z, Liu B, Zheng X, Zhang D, Malzahn A, Gong Z, Qi Y, Zhang T, Zhang Y (2018) A large-scale whole-genome sequencing analysis reveals highly specific genome editing by Both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol 19:84
Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y (2017) A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nature Plant 3:17018. https://doi.org/10.1038/nplants.2017.18
Terns MP, Terns RM (2011) CRISPR-based adaptive immune systems. Curr Opin Microbiol 14:321–327
Veillet F, Kermarrec MP, Chauvin L, Guyon-Debast A, Chauvin JE, Gallois JL, Nogué F (2020) Prime editing is achievable in the tetraploid potato, but needs improvement. bioRxiv. https://doi.org/10.1101/2020.06.18.159111
von Arnim AG, Jia Q, Vaughn JN (2014) Regulation of plant translation by upstream open reading frames. Plant Science 214:1–12
Voytas DF (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64:327–350
Wada N, Ueta R, Osakabe Y, Osakabe K (2020) Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol 20:234. https://doi.org/10.1186/s12870-020-02385-5
Wang L, Kaya HB, Zhang N, Rai R, Willmann MR, Carpenter SCD, Read AC, Martin F, Fei Z, Leach JE, Martin GB, Bogdanove AJ (2021) Spelling changes and fluorescent tagging with prime editing vectors for plants. Front Genome Edit 3:7. https://doi.org/10.3389/fgeed.2021.617553
Wang M, Lu Y, Botella JR, Mao Y, Hua K, Zhu JK (2017) Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol Plant 10:1007–1010
Woo JW, Kim J, Kwon SI, Corvalán C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164
Wright DA, Townsend JA, Winfrey RJ Jr, Irwin RA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF (2005) High-frequency homologous recombination in plants mediated by zinc-finger nucleases: recombination and zinc-finger nucleases. Plant J 44:693–705
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci 112:3570–3575
Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J (2017) Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J 15:713–717
Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Markavo KS, Koonin EV, Ishitani R, Zhang F, Nureki O (2016) Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165:949–962
Yin X, Biswal AK, Dionora J, Perdigon KM, Balahadia CP, Mazumdar S, Chater C, Lin H-C, Coe RA, Kretzschmar T, Gray JE, Quick PW, Bandypodhyay A (2017) CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep 36:745–757
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771
Zhang D, Zhang H, Li T, Chen K, Qiu JL, Gao C (2017a) Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol 18:191. https://doi.org/10.1186/s13059-017-1325-9
Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P (2011) Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29:149–153
Zhang H, Si X, Ji X, Fan R, Liu J, Chen K, Wang D, Gao C (2018) Genome editing of upstream open reading frames enables translational control in plants. Nat Biotechnol 36:894–898
Zhang X, Wang J, Cheng Q, Zheng X, Zhao G, Wang J (2017b) Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov 3:17018. https://doi.org/10.1038/celldisc.2017.18
Zhang Y, Malzahn AA, Sretenovic S, Qi Y (2019) The emerging and uncultivated potential of CRISPR technology in plant science. Nature Plant 5:778–794
Zhong Z, Sretenovic S, Ren Q, Yang L, Bao Y, Qi C, Yuan M, He Y, Liu S, Liu X, Wang J, Huang L, Wang Y, Baby D, Wang D, Zhang T, Qi Y, Zhang Y (2019) Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol Plant 12:1027–1036
Zhong Z, Zhang Y, You Q, Tang X, Ren Q, Liu S, Yang L, Wang Y, Liu Z, Liu B, Zhang T, Zheng X, Le Y, Zhang Y, Qi Y (2018) Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites. Mol Plant 11:999–1002
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nuc Acid Res 42:10903–10914
Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol 36:1211–1216. https://doi.org/10.1038/nbt.4272
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: Gary Bannon
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zess, E., Begemann, M. CRISPR-Cas9 and beyond: what’s next in plant genome engineering. In Vitro Cell.Dev.Biol.-Plant 57, 584–594 (2021). https://doi.org/10.1007/s11627-021-10185-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-021-10185-1