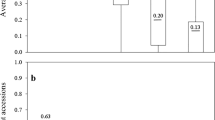
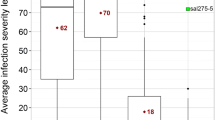
Abstract
Bremia lactucae is a devastating pathogen causing heavy yield losses in lettuce production worldwide. Long time it was thought to infect various members of the Asteraceae but recent phylogenetic investigations revealed that B. lactucae is restricted to cultivated Lactuca sativa and its wild progenitor Lactuca serriola. Many resistance genes in cultivated lettuce were crossbred from L. serriola, but often a fast overcoming of the resistance in the crop by new downy mildew races was observed. Consequently, it seems important to investigate the population genetic relationships of wild prickly lettuce and cultivated lettuce, in order to determine, if the Bremia lineages infecting these hosts are distinct or intermingled. In the first case, searching for resistance in prickly lettuce is straightforward, in the second case it would rather be futile. Bremia belongs to the genera of downy mildews with pyriform haustoria, which feature highly variable ITS-repeats that can be used as high-resolution markers for population genetics studies. Here we report that Bremia lactucae on Lactuca sativa is not homogeneous but emerged from several host shifts from Lactuca serriola and that Bremia accessions from both species are not forming distinct genepools. While the pathogen population from L. serriola showed a balanced pattern, the pathogen population from L. sativa has a directional selection pattern reflecting founder effects after host shifts, resulting in a fast colonisation of susceptible lettuce cultivars. Thus, the wild population of Bremia lactucae seems to play a major role in the epidemiology of downy mildew on cultivated lettuce. This also explains, why resistance breeding using resistance genes from prickly lettuce had little success for achieving durable resistance in cultivated lettuce and calls for both more detailed research into Bremia on wild lettuce and new breeding strategies using less closely related lettuce species.


Similar content being viewed by others
Change history
31 December 2021
A Correction to this paper has been published: https://doi.org/10.1007/s10658-021-02338-0
References
Bachofer, M. (2004). Molekularbiologische Populationsstudien an Plasmopara halstedii dem Falschen Mehltau der Sonnenblume, Dissertation Universität Hohenheim, , 1–140.
Barrière, V., Lecompte, F., Nicot, P. C., Maisonneuve, B., Tchamitchian, M., & Lescourret, F. (2014). Lettuce cropping with less pesticides. A review. Agronomy for Sustainable Development, 34, 175–198.
Beharav, A., Ochoa, O., & Michelmore, R. (2014). Resistance in natural populations of three wild Lactuca species from Israel to highly virulent Californian isolates of Bremia lactucae. Genetic Resources and Crop Evolution, 61, 603–609.
Bohn, G. W., & Whitaker, T. W. (1951). Recently introduced varieties of head lettuce and methods used in their development. US Department of Agriculture Circular, 881.
Choi, Y. J., & Thines, M. (2015). Host jumps and radiation, not co-divergence drives diversification of obligate pathogens. A case study in downy mildews and Asteraceae. PloS ONE, 10(7), e0133655.
Choi, Y.-J., Hong, S.-B., & Shin, H.-D. (2007). Extreme size and sequence variation in the ITS rDNA of Bremia lactucae. Mycopathologia, 163, 91–95.
Choi, Y.-J., Thines, M., Runge, F., Hong, S.-B., Telle, S., & Shin, H.-D. (2011). Evidence for high degrees of specialisation, evolutionary diversity, and morphological distinctiveness in the genus Bremia. Fungal Biology, 115, 102–111.
Choi, Y. J., Wong, J., Runge, F., Mishra, B., Michelmore, R., & Thines, M. (2017). BrRxLR11 – A new phylogenetic marker with high resolution in the downy mildew genus Bremia and related genera. Mycological Progress, 16, 185–190.
Cooke, D. E. L., Drenth, A., Duncan, J. M., Wagels, G., & Brasier, C. M. (2000). A molecular phylogeny of phytophthora and related oomycetes. Fungal Genetics and Biology, 30, 17–32.
Cousens, R., & Croft, A. M. (2000). Weed populations and pathogens. Weed Research, 40, 63–82.
Crute, I. R. (1981). The host specificity of peronosporaceous fungi and the genetics of the relationship between host and parasite. In D. M. Spencer (Ed.), The downy mildews (pp. 237–253). Academic Press.
Crute, I. R. (1992). From breeding to cloning (and back again?): A case study with lettuce downy mildew. Annual Review of Phytopathology, 30, 485–506.
Crute, I. R., & Johnson, A. G. (1976). The genetic relationship between races of Bremia lactucae and cultivars of Lactuca sativa. Annals of Applied Biology, 83, 125–137.
D’Andrea, L., Felber, F., & Guadagnuolo, R. (2008). Hybridization rates between lettuce (Lactuca sativa) and its wild relative (L. serriola) under field conditions. Environmental Biosafety Research, 7, 61–71.
de Vries, I. M. (1997). Origin and domestication of Lactuca sativa L. Genetic Resources and Crop Evolution, 44, 165–174.
Excoffier, L., & Lischer, H. E. L. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and windows. Molecular Ecology Resources, 10, 564–567.
Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131, 479–491.
FAOSTAT (2018). http://www.fao.org/faostat/en/#data/QV. Accessed 12 Dec 2018.
Farrara, B. F., Illot, T. W., & Michelmore, R. W. (1987). Genetic analysis of factors for resistance to downy mildew (Bremia lactucae) in species of lettuce (Lactuca sativa and L. serriola). Plant Pathology, 36, 499–514.
Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39, 783–791.
Gärber, U., & Idczak, E. (2007). Erhebungen zum Auftreten und zum Rassenspektrum von Bremia lactucae an Salat in Deutschland. Nachrichtenblatt des Deutschen Pflanzenschutzdienstes, 59(10), 221–226.
Huelsenbeck, J. P., & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Hulbert, S. H., & Michelmore, R. W. (1985). Linkage analysis of genes for resistance to downy mildew (Bremia lactucae) in lettuce (Lactuca sativa). Theoretical and Applied Genetics, 70, 520–528.
Ito, S., & Tokunaga, Y. (1935). Notae mycologicae. Asiae orientalis I. Transaction of the Sapporo Natural History Society, 14, 11–33.
Jemelková, M., Kitner, M., Křístková, E., Beharav, A., & Lebeda, A. (2015). Biodiversity of Lactuca aculeata germplasm assessed by SSR and AFLP markers, and resistance variation to Bremia lactucae. Biochemical Systematics and Ecology, 61, 344–356.
Jones, J. D., & Dangl, J. L. (2006). The plant immune system. Nature, 444(7117), 323–329.
Katoh, K., Kuma, K.-I., Toh, H., & Miyata, T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33, 511–518.
Kesseli, R. V., Ochoa, O., & Michelmore, R. W. (1991). Variation at RFLP loci in Lactuca spp. and the origin of cultivated lettuce (L. sativa). Genome, 34, 430–436.
Kitner, M., Majeský, L., Křístková, E., Jemelková, M., Lebeda, A., & Beharav, A. (2015). Genetic structure and diversity in natural populations of three predominantly self-pollinating wild Lactuca species in Israel. Genetic Resources and Crop Evolution, 62, 991–1008.
Kroon, L. P. N. M., Bakker, F. T., Van den Bosch, G. B. M., Bonants, P. J. M., & Flier, W. G. (2004). Phylogenetic analysis of Phytophthora species based on mitochondrial andnuclear DNA sequences. Fungal Genetics and Biology, 41, 766–782.
Lebeda, A. (1989). Response of lettuce cultivars carrying the resistance gene Dm11 to isolates of Bremia lactucae from Lactuca serriola. Plant Breeding, 102, 311–316.
Lebeda, A. (2002). Occurrence and variation in virulence of Bremia lactucae in natural populations of Lactuca serriola. In P. T. N. Spencer Phillips, U. Gisi, & A. Lebeda (Eds.), Advances in downy mildew research (pp. 179–183). Kluwer Academic Publishers.
Lebeda, A., & Blok, I. (1990). Sexual compatibility types of Bremia lactucae isolates originating from Lactuca serriola. Netherlands Journal of Plant Pathology, 96, 51–54.
Lebeda, A., & Petrželová, I. (2004a). Variation and distribution of virulence phenotypes of Bremia lactucae in natural populations of Lactuca serriola. Plant Pathology, 53, 316–324.
Lebeda, A., & Petrželová, I. (2004b). Occurrence of race-specific resistance to Bremia lactucae in Lactuca serriola germplasm originating from European countries. In J. Vollmann, H. Grausgruber, & P. Ruckenbauer (Eds.), Genetic variation for plant breeding (pp. 113–116). EUCARPIA & BOKU-University of Natural Resources and Applied Life Sciences.
Lebeda, A., & Syrovátko, P. (1988). Specificity of Bremia lactucae isolates from Lactuca sativa and some Asteraceae plants. Acta Phytopathologica et Entomologica Hungarica, 23, 39–48.
Lebeda, A., & Zinkernagel, V. (2003a). Evolution and distribution of virulence in the German population of Bremia lactucae. Plant Pathology, 52, 41–51.
Lebeda, A., & Zinkernagel, V. (2003b). Characterization of new highly virulent German isolates of Bremia lactucae and efficiency of resistance in wild Lactuca spp. germplasm. Journal of Phytopathology, 151, 274–282.
Lebeda, A., Doležalová, I., Křístková, E., & Mieslerová, B. (2001). Biodiversity and ecogeography of wild Lactuca spp. in some European countries. Genetic Resources and Crop Evolution, 48, 153–164.
Lebeda, A., Pink, D. A. C., & Astley, D. (2002). Aspects of the interactions between wild Lactuca spp. and related genera and lettuce downy mildew (Bremia lactucae). In P. T. N. Spencer Phillips, U. Gisi, & A. Lebeda (Eds.), Advances in downy mildew research (pp. 85–117). Kluwer Academic Publishers.
Lebeda, A., Ryder, E. J., Grube, R., Doležalová, I., & Křístková, E. (2007a). Lettuce (Asteraceae; Lactuca spp.), chapter 9. In R. Singh (Ed.), Genetic resources, chromosome engineering, and crop improvement series, volume 3 – Vegetable crops (pp. 377–472). CRC.
Lebeda, A., Petrželová, I., & Maryška, Z. (2007b). Comparative analysis of variation and dynamics of virulence of Bremia lactucae populations on Lactuca sativa and Lactuca serriola. In B. MacDonald, P. Brunner, P Ceresini & A. Lebeda (Eds.), Population and evolutionary biology of fungal symbionts (pp. 50–51). Book of Abstracts, International Meeting, April 29–May 4, 2007, Ascona, Switzerland; ETHZ-Swiss Federal Institute of Technology of Zurich and Palacký University in Olomouc; Zurich and Olomouc. JOLA, v.o.s., Kostelec na Hané, Czech Republic.
Lebeda, A., Petrželová, I., & Maryška, Z. (2008a). Structure and variation in the wild-plant pathosystem: Lactuca serriola - Bremia lactucae. European Journal of Plant Pathology, 122, 127–146.
Lebeda, A., Sedlářová, M., Petřivalský, M., & Prokopová, J. (2008b). Diversity of defence mechanisms in plant-oomycete interactions: A case study of Lactuca spp. and Bremia lactucae. European Journal of Plant Pathology, 122, 71–89.
Lebeda, A., Křístková, E., Kitner, M., Mieslerová, B., Jemelková, M., & Pink, D. A. C. (2014). Wild Lactuca species, their genetic diversity, resistance to diseases and pests, and exploitation in lettuce breeding. European Journal of Plant Pathology, 138, 597–640.
Librado, P., & Rozas, J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11), 1451–1452.
Lindqvist, K. (1960). On the origin of cultivated lettuce. Hereditas, 46, 319–350.
Michelmore, R. W., &Wong, J. (2008). Classical and molecular genetics of Bremia lactucae, cause of lettuce downy mildew. European Journal of Plant Pathology, 122, 19–30.
Michelmore, R. W., Ochoa, O. E., &Wong, J. (2009). Bremia lactucae and lettuce downy mildew. In K. Lamour &S. Kamoun (Eds.), Oomycete genetics and genomics: Diversity, interactions, and research tools (pp. 241–262). J. Wiley.
Mishra, B., & Thines, M. (2014). siMBa—A simple graphical user interface for the Bayesian phylogenetic inference program MrBayes. Mycological Progress, 13, 1255–1258.
Moncalvo, J. M., Wang, H. H., & Hseu, R. S. (1995). Phylogenetic relationships in Ganoderma inferred from the internal transcribed spacers and 25S ribosomal DNA sequences. Mycologia, 87, 223–238.
Nordskog, B., Elameen, A., Gadoury, D. M., & Hermansen, A. (2014). Virulence characteristics of Bremia lactucae populations in Norway. European Journal of Plant Pathology, 139, 679–686.
Park, J. H., Thines, M., Lee, H. B., Shin, H.-D., & Choi, Y.-J. (2018). Bremia polycephala and Bremia sawadae spp. nov. (Peronosporaceae; Oomycota), parasitic to northeast Asian Asteraceae. Nova Hedwegia, 107, 303–317.
Parra, L., Maisonneuve, B., Lebeda, A., Schut, J., Christopoulou, M., Jeuken, M., McHale, L., Truco, M.-J., Crute, I. R., & Michelmore, R. (2016). Rationalization of genes for resistance to Bremia lactucae in lettuce. Euphytica, 210, 309–326.
Petrželová, I., & Lebeda, A. (2003). Distribution of compatibility types and occurrence of sexual reproduction in natural populations of Bremia lactucae on wild Lactuca serriola plants. Acta Phytopathologica et Entomologica Hungarica, 38, 43–52.
Petrželová, I., & Lebeda, A. (2004). Temporal and spatial variation in virulence of natural populations of Bremia lactucae occurring on Lactuca serriola. In P. T. N. Spencer Phillips &M. Jeger (Eds.), Advances in downy mildew research (Vol. 2, pp. 141–163). Kluwer Academic Publishers.
Petrželová, I., & Lebeda, A. (2011). Distribution of race-specific resistance against Bremia lactucae in natural populations of Lactuca serriola. European Journal of Plant Pathology, 129, 233–253.
Petrželová, I., Lebeda, A., & Kosman, E. (2013). Distribution, disease level and virulence variation of Bremia lactucae on Lactuca sativa in the Czech Republic in the period 1999-2011. Journal of Phytopathology, 161, 503–514.
Regel, E. (1843). Beiträge zur Kenntnis einiger Blattpilze. Botanische Zeitung, 1, 665–667.
Runge, F., & Thines, M. (2009). A potential perennial host for Pseudoperonospora cubensis in temperate regions. European Journal of Plant Pathology, 123, 483–486.
Sain, S. K., Monga, D., & Dewasi, H. (2018). First report of downy mildew caused by Bremia lactucae on Sonchus asper L. (hill) and S. tenerrimus L. in Haryana, India. Indian Phytopathology, 71, 465–467.
Sawada, K. (1914). Notes on the species of Bremia. The Botanical Magazine, 28, 132–140.
Sharaf, K., Lewinsohn, D., Nevo, E., &Beharav, A. (2007). Virulence patterns of Bremia lactucae in Israel. Phytoparasitica, 35, 100–108.
Skidmore, D. I., & Ingram, D. S. (1985). Conidial morphology and the specialization of Bremia lactucae Regel (Peronosporaceae) on hosts in the family Compositae. Botanical Journal of the Linnean Society, 91, 503–522.
Spring, O., Gomez-Zeledon, J., Hadziabdic, D., Trigiano, R. N., Thines, M., & Lebeda, A. (2018). Biological characteristics and assessment of virulence diversity in pathosystems of economically important biotrophic oomycetes. Critical Reviews in Plant Sciences, 37, 439–495.
Stamatakis, A., Hoover, P., & Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web-servers. Systematic Biology, 75, 758–771.
Stukenbrock, E. H., Quaedvlieg, W., Javan-Nikhah, M., Zala, M., Crous, P. W., & McDonald, B. A. (2012). Zymoseptoria ardabiliae and Z. pseudotritici, two progenitor species of the septoria tritici leaf blotch fungus Z. tritici (synonym: Mycosphaerella graminicola). Mycologia, 104(6), 1397–1407.
Sydow, H. (1923). Mycotheca germanica Fasc. 37-41 (Nrs. 1801-2050). Annals of Mycology, 21, 165–181.
Sydow, H. (1938). Mycotheca germanica Fasc. 61-64 (Nrs. 3001-3200). Annals of Mycology, 26, 318–325.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evololution, 30, 2725–2729.
Tao, J.-F., & Yu, Y.-N. (1992). Taxa of the genus Bremia on the family Compositae from China. Acta Mycologica Sinica, 11, 89–95.
Teacher, A. G. F., & Griffiths, D. J. (2010). HapStar: Automated haplotype network layout and visualization. Molecular Ecology Resources, online.
Telle, S., & Thines, M. (2008). Amplification of cox2 (~620bp) from 2 mg of up to 129 years old herbarium specimens, comparing 19 extraction methods and 15 polymerases. PLoS One, 3, e3584.
Thines, M. (2007). Characterisation and phylogeny of repeated elements giving rise to exceptional length of ITS2 in several downy mildew genera (Peronosporaceae). Fungal Genetics and Biology, 44, 199–207.
Thines, M. (2019). An evolutionary framework for host shifts–jumping ships for survival. New Phytologist, 224(2), 605–617.
Thines, M., Runge, F., Telle, S., & Voglmayr, H. (2010). Phylogenetic investigations in the downy mildew genus Bremia reveal several distinct lineages and a species with a presumably exceptional wide host range. European Journal of Plant Pathology, 128, 81–89.
Van Hese, N., Huang, C. J., De Vleesschauwer, D., Delaere, I., Pauwelyn, E., Bleyaert, P., &Höfte, M. (2016). Evolution and distribution of virulence characteristics of Belgian Bremia lactucae populations between 2008 and 2013. European Journal of Plant Pathology, 144, 431–441.
Van Treuren, R., & van Hintum, T. J. L. (2009). Comparison of anonymous and targeted molecular markers for the estimation of genetic diversity in ex situ conserved Lactuca. Theoretical and Applied Genetics, 119, 1265–1279.
van Treuren, R., van Hintum, T. J. L., & van de Wiel, C. C. M. (2008). Marker-assisted optimization of an expert-based strategy for the acquisition of modern lettuce varieties to improve a genebank collection. Genetic Resources and Crop Evolution, 55, 319–330.
van Treuren, R., Van der Arend, A., & Schut, J. (2013). Distribution of downy mildew (Bremia lactucae Regel) resistances in a genebank collection of lettuce and its wild relatives. Plant Genetic Resources: Characterisation and Utilisation, 11, 15–25.
Vieira, B. S., & Barretto, R. W. (2006). First record of Bremia lactucae infecting Sonchus oleraceus and Sonchus asper in Brazil and its infectivity to lettuce. Journal of Phytopathology, 154, 84–87.
Voglmayr, H., & Constantinescu, O. (2008). Revision and reclassification of three Plasmopara species based on morphological and molecular phylogenetic data. Mycological Research, 112, 487–501.
Voglmayr, H., Riethmüller, A., Göker, M., Weiß, M., & Oberwinkler, F. (2004). Phylogenetic relationships of Plasmopara, Bremia and other genera of downy mildews with pyriform haustoria based on Bayesian analysis of partial LSU rDNA sequence data. Mycological Research, 108, 1011–1024.
Wright, S. (1965). The interpretation of population structure by F-statistics with special regards to systems of mating. Evolution, 19, 395–420.
Yerkes, W. D., & Shaw, C. G. (1959). Taxonomy of Peronospora species on Cruciferae and Chenopodiaceae. Phytopathology, 49, 499–507.
Zohary, D. (1991). The wild genetic resources of cultivated lettuce (Lactuca sativa L.). Euphytica., 53, 31–35.
Funding
This study was supported by a grant from the Ministry of Science, Education, and the Arts of Baden-Württemberg awarded to F. Runge and by the research funding program “LOEWE—Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz” of Hesse’s Ministry of Higher Education, Research, and the Arts in the framework of the Centre for Translational Biodiversity Genomics (TBG) and the Cluster for Integrative Fungal Research (IPF). A. Lebeda acknowledge the support of grant MSM 6198959215 (funded by Ministry of Education, Youth and Sports of the Czech Republic), the National Programme of Genepool Conservation of Microorganisms and Small Animals of Economic Importance of the Czech Republic, and Internal Grant Agency of Palacký University in Olomouc (IGA_PrF_2018_001; IGA_PrF_2019_004; IGA_PrF_2020 003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The Authors declare that the present manuscript complies with the Ethical Rules of good scientific practice applicable for the European Journal of Plant Pathology.
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
No human and/or animal participants were involved in this research.
Informed consent
All authors consent to this submission.
Rights and permissions
About this article
Cite this article
Runge, F., Gärber, U., Lebeda, A. et al. Bremia lactucae populations on cultivated lettuce originate from prickly lettuce and are interconnected with the wild pathosystem. Eur J Plant Pathol 161, 411–426 (2021). https://doi.org/10.1007/s10658-021-02332-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-021-02332-6