Abstract
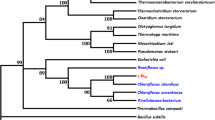
An alkaline esterase, designated as EstXT1, was identified through functional screening from a metagenomic library. Sequence analysis revealed that EstXT1 belonged to the family VIII carboxylesterases and contained a characteristic conserved S-x-x-K motif and a deduced catalytic triad Ser56-Lys59-Tyr165. EstXT1 exhibited the strongest activity toward methyl ferulate at pH 8.0 and temperature 55°C and retained over 80% of its original activity after incubation in the pH range of 7.0–10.6 buffers. Biochemical characterization of the recombinant enzyme showed that it was activated by Zn2+ and Co2+ metal ion, while inhibited by Cu2+ and CTAB. EstXT1 exhibited significant promiscuous acyltransferase activity preferred to the acylation of benzyl alcohol acceptor using short-chain pNP-esters (C2-C8) as acyl-donors. A structure–function analysis indicated that a WAG motif is essential to acyltransferase activity. This is the first report example that WAG motif plays a pivotal role in acyltransferase activity in family VIII carboxylesterases beside WGG motif. Further experiment indicated that EstXT1 successfully acylated cyanidin-3-O-glucoside in aqueous solution. The results from the current investigation provided new insights for the family VIII carboxylesterase and lay a foundation for the potential applications of EstXT1 in food and biotechnology fields.








Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article.
References
Arpigny, J. L., & Jaeger, K.-E. (1999). Bacterial lipolytic enzymes: classification and properties. The Biochemical Journal, 343(1), 177.
Ramnath, L., Sithole, B., & Govinden, R. (2017). Classification of lipolytic enzymes and their biotechnological applications in the pulping industry. Canadian Journal of Microbiology, 63(3), 179–192.
Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., & Goldman, A. (1992). The α/β hydrolase fold. Protein Engineering, 5(3), 197–211.
Kovacic, F., Babic, N., Krauss, U., & Jaeger, K.-E. (2018). Classification of lipolytic enzymes from bacteria. In F. Rojo (Ed.), Aerobic utilization of hydrocarbons, oils and lipids (pp. 1–35). Springer International Publishing.
Ma, F., Chung, M. T., Yao, Y., Nidetz, R., Lee, L. M., Liu, A. P., Feng, Y., Kurabayashi, K., & Yang, G.-Y. (2018). Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform. Nature Communications, 9, 1–8.
Ramya, S. L., Venkatesan, T., Murthy, K. S., Jalali, S. K., & Verghese, A. (2016). Detection of carboxylesterase and esterase activity in culturable gut bacterial flora isolated from diamondback moth, Plutella xylostella (Linnaeus), from India and its possible role in indoxacarb degradation. Brazilian Journal of Microbiology, 47(2), 327–336.
Bornscheuer, U. T. (2002). Microbial carboxyl esterases: classification, properties and application in biocatalysis. Fems Microbiol. Rev, 26(1), 73–81.
Hitch, T. C. A., & Clavel, T. (2019). A proposed update for the classification and description of bacterial lipolytic enzymes. PeerJ, 7, e7249.
Verma, N., Thakur, S., & Bhatt, A. K. (2012). Microbial lipases: industrial applications and properties. International Journal of Biological Sciences, 1, 88–92.
Riesenfeld, C. S., Schloss, P. D., & Handelsman, J. (2004). Metagenomics: genomic analysis of microbial communities. Annual Review of Genetics, 38(1), 525–552.
Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J., & Goodman, R. M. (1998). Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemistry & Biology, 5(10), 245–249.
Steele, H. L., Jaeger, K. E., Daniel, R., & Streit, W. R. (2009). Advances in recovery of novel biocatalysts from metagenomes. Journal of Molecular Microbiology and Biotechnology, 16(1-2), 25–37.
Milshteyn, A., Schneider, J. S., & Brady, S. F. (2014). Mining the metabiome: identifying novel natural products from microbial communities. Chemistry & Biology, 21(9), 1211–1223.
Castillo Villamizar, G. A., Nacke, H., Boehning, M., Herz, K., Daniel, R., & Barkan, D. (2019). Functional metagenomics reveals an overlooked diversity and novel features of soil-derived bacterial phosphatases and phytases. mBio, 10(1) e01966-18.
Sarkar, J., Dutta, A., Chowdhury, P. P., Chakraborty, J., & Dutta, T. K. (2020). Characterization of a novel family VIII esterase EstM2 from soil metagenome capable of hydrolyzing estrogenic phthalates. Microbial Cell Factories, 19(1), 77–77.
Kang, H.-S., & Brady, S. F. (2013). Arimetamycin A: Improving clinically relevant families of natural products through sequence-guided screening of soil metagenomes. Angewandte Chemie, International Edition, 52(42), 11063–11067.
Tchigvintsev, A., Tran, H., Popovic, A., Kovacic, F., Brown, G., Flick, R., Hajighasemi, M., Egorova, O., Somody, J. C., Tchigvintsev, D., Khusnutdinova, A., & Chernikova, T. N. (2015). The environment shapes microbial enzymes: five cold-active and salt-resistant carboxylesterases from marine metagenomes. Applied Microbiology and Biotechnology, 99(5), 2165–2178.
Zhang, Y., Hao, J., Zhang, Y.-Q., Chen, X.-L., Xie, B.-B., Shi, M., Zhou, B.-C., Zhang, Y.-Z., & Li, P.-Y. (2017). Identification and characterization of a novel salt-tolerant esterase from the deep-sea sediment of the South China Sea. Frontiers in Microbiology, 8(441), 1–8.
Lu, M., Dukunde, A., & Daniel, R. (2019). Biochemical profiles of two thermostable and organic solvent-tolerant esterases derived from a compost metagenome. Applied Microbiology and Biotechnology, 103(8), 3421–3437.
Ranjan, R., Yadav, M. K., Suneja, G., & Sharma, R. (2018). Discovery of a diverse set of esterases from hot spring microbial mat and sea sediment metagenomes. International Journal of Biological Macromolecules, 119, 572–581.
Gupta, V., Singh, I., Kumar, P., Rasool, S., & Verma, V. (2019). A hydrolase with esterase activity expressed from a fosmid gene bank prepared from DNA of a North West Himalayan glacier frozen soil sample. 3. Biotech, 9(3), 1071–1077.
De Santi, C., Ambrosino, L., Tedesco, P., Zhai, L., Zhou, C., Xue, Y., Ma, Y., & de Pascale, D. (2015). Identification and characterization of a novel salt-tolerant esterase from a Tibetan glacier metagenomic library. Biotechnology Progress, 31(4), 890–899.
Heath, C., Hu, X. P., Cary, S. C., & Cowan, D. (2009). Identification of a novel alkaliphilic esterase active at low temperatures by screening a metagenomic library from Antarctic desert soil. Applied and Environmental Microbiology, 75(13), 4657–4659.
Kim, Y.-O., Park, I.-S., Nam, B.-H., Kim, D.-G., Jee, Y.-J., Lee, S.-J., & An, C.-M. (2014). A novel esterase from Paenibacillus sp PBS-2 is a new member of the beta-lactamase belonging to the family VIII lipases/esterases. Journal of Microbiology and Biotechnology, 24(9), 1260–1268.
Zhang, H., Li, M., Dai, C., Wang, G., Xiong, M., Li, F., Liu, Y., & Xu, D. (2016). Characterization of EstQE, a new member of esterase family VIII from the quizalofop-P-ethyl-degrading bacterium Ochrobactrum sp QE-9. Journal of Molecular Catalysis B: Enzymatic, 133, 167–175.
Jeon, J. H., Lee, H. S., Lee, J. H., Koo, B.-S., Lee, C.-M., Lee, S. H., Kang, S. G., & Lee, J.-H. (2016). A novel family VIII carboxylesterase hydrolysing third- and fourth-generation cephalosporins. SpringerPlus, 5(1), 525–525.
Brady, S. F. (2007). Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nature Protocols, 2(5), 1297–1305.
Hancock, J., MZvelebil, M. J., (2004) Dictionary of bioinformatics and computational biology. Wiley-Liss.
Altschul, S. F., Wootton, J. C., Gertz, E. M., Agarwala, R., Morgulis, A., Schaffer, A. A., & Yu, Y. K. (2005). Protein database searches using compositionally adjusted substitution matrices. The FEBS Journal, 272(20), 5101–5109.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., & Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23(21), 2947–2948.
Robert, X., & Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42(Web Server issue) W320-4.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549.
Mueller, H., Godehard, S. P., Palm, G. J., Berndt, L., Badenhorst, C. P. S., Becker, A.-K., Lammers, M., & Bornscheuer, U. T. (2021). Discovery and design of family VIII carboxylesterases as highly efficient acyltransferases. Angewandte Chemie, International Edition, 60(4), 2013–2017.
Reisky, L., Srinivasamurthy, V. S. T., Badenhorst, C. P. S., Godehard, S. P., & Bornscheuer, U. T. (2019). A novel high-throughput assay enables the direct identification of acyltransferases. Catalysts, 9(1).
Majiduddin, F. K., Materon, I. C., Palzkill, T. G., (2002) Molecular analysis of beta-lactamase structure and function. International Journal of Medical Microbiology 292 (2), 0-137, 127.
Pratt, R. F. (2008). Substrate specificity of bacterial DD-peptidases (penicillin-binding proteins). Cellular and Molecular Life Sciences, 65(14), 2138–2155.
Cha, S. S., An, Y. J., Jeong, C. S., Kim, M. K., Jeon, J. H., Lee, C. M., Lee, H. S., Kang, S. G., & Lee, J. H. (2013). Structural basis for the beta-lactamase activity of EstU1, a family VIII carboxylesterase. Proteins, 81(11), 2045–2051.
Jeon, J. H., Kim, S. J., Lee, H. S., Cha, S. S., Lee, J. H., Yoon, S. H., Koo, B. S., Lee, C. M., Choi, S. H., Lee, S. H., Kang, S. G., & Lee, J. H. (2011). Novel metagenome-derived carboxylesterase that hydrolyzes beta-lactam antibiotics. Applied and Environmental Microbiology, 77(21), 7830–7836.
Rashamuse, K., Magomani, V., Ronneburg, T., & Brady, D. (2009). A novel family VIII carboxylesterase derived from a leachate metagenome library exhibits promiscuous β-lactamase activity on nitrocefin. Applied Microbiology and Biotechnology, 83(3), 491–500.
Mokoena, N., Mathiba, K., Tsekoa, T., Steenkamp, P., & Rashamuse, K. (2013). Functional characterisation of a metagenome derived family VIII esterase with a deacetylation activity on beta-lactam antibiotics. Biochemical and Biophysical Research Communications, 437(3), 342–348.
Biver, S., & Vandenbol, M. (2013). Characterization of three new carboxylic ester hydrolases isolated by functional screening of a forest soil metagenomic library. Journal of Industrial Microbiology & Biotechnology, 40(2), 191–200.
Lee, H. W., Jung, W. K., Kim, Y. H., Ryu, B. H., Kim, T. D., Kim, J., & Kim, H. (2016). Characterization of a novel alkaline family VIII esterase with S-enantiomer preference from a compost metagenomic library. Journal of Industrial Microbiology & Biotechnology, 26(2), 315–325.
Wu, S., Nan, F., Jiang, J., Qiu, J., Zhang, Y., Qiao, B., Li, S., & Xin, Z. (2019). Molecular cloning, expression and characterization of a novel feruloyl esterase from a soil metagenomic library with phthalate-degrading activity. Biotechnology Letters, 41(8-9), 995–1006.
Cruz, L., Fernandes, I., Guimaraes, M., de Freitas, V., & Mateus, N. (2016). Enzymatic synthesis, structural characterization and antioxidant capacity assessment of a new lipophilic malvidin-3-glucoside-oleic acid conjugate. Food & Function, 7(6), 2754–2762.
Cruz, L., Benohoud, M., Rayner, C. M., Mateus, N., de Freitas, V., & Blackburn, R. S. (2018). Selective enzymatic lipophilization of anthocyanin glucosides from blackcurrant (Ribes nigrum L.) skin extract and characterization of esterified anthocyanins. Food Chemistry, 266, 415–419.
Yang, W., Kortesniemi, M., Ma, X., Zheng, J., & Yang, B. (2019). Enzymatic acylation of blackcurrant (Ribes nigrum) anthocyanins and evaluation of lipophilic properties and antioxidant capacity of derivatives. Food Chemistry, 281, 189–196.
Park, J.-M., Won, S.-M., Kang, C.-H., Park, S., & Yoon, J.-H. (2020). Characterization of a novel carboxylesterase belonging to family VIII hydrolyzing beta-lactam antibiotics from a compost metagenomic library. International Journal of Biological Macromolecules, 164, 4650–4661.
Mueller, H., Godehard, S. P., Palm, G. J., Berndt, L., Badenhorst, C. P. S., Becker, A.-K., Lammers, M., Bornscheuer, U. T., (2020) Discovery and design of family VIII carboxylesterases as highly efficient acyltransferases. Angew. Chem.-Int. Edit..
Silva, S., Costa, E. M., Mendes, M., Morais, R. M., Calhau, C., & Pintado, M. M. (2016). Antimicrobial, antiadhesive and antibiofilm activity of an ethanolic, anthocyanin-rich blueberry extract purified by solid phase extraction. Journal of Applied Microbiology, 121(3), 693–703.
Ruenroengklin, N., Yang, B., Lin, H., Chen, F., & Jiang, Y. (2009). Degradation of anthocyanin from litchi fruit pericarp by H2O2 and hydroxyl radical. Food Chemistry, 116(4), 995–998.
Yang, W., Kortesniemi, M., Yang, B., & Zheng, J. (2018). Enzymatic acylation of anthocyanins isolated from alpine bearberry (Arctostaphylos alpina) and lipophilic properties, thermostability, and antioxidant capacity of the derivatives. Journal of Agricultural and Food Chemistry, 66(11), 2909–2916.
Kazemi, M., Sheng, X., Kroutil, W., & Himo, F. (2018). Computational study of Mycobacterium smegmatis acyl transferase reaction mechanism and specificity. ACS Catalysis, 8(11), 10698–10706.
Mathews, I., Soltis, M., Saldajeno, M., Ganshaw, G., Sala, R., Weyler, W., Cervin, M. A., Whited, G., & Bott, R. (2007). Structure of a novel enzyme that catalyzes acyl transfer to alcohols in aqueous conditions. Biochemistry, 46(31), 8969–8979.
Jost, E., Kazemi, M., Mrkonjic, V., Himo, F., Winkler, C. K., & Kroutil, W. (2020). Variants of the acyltransferase from Mycobacterium smegmatis enable enantioselective acyl transfer in water. ACS Catalysis, 10(18), 10500–10507.
Godehard, S. P., Badenhorst, C. P. S., Mueller, H., & Bornscheuer, U. T. (2020). Protein engineering for enhanced acyltransferase activity, substrate scope, and selectivity of the Mycobacterium smegmatis acyltransferase MsAcT. ACS Catalysis, 10(14), 7552–7562.
Funding
This research was supported by the Key Research and Development Program of Jiangsu Province (BE2017374-2) and special funds of agroproduct quality safety risk assessment of the Ministry of Agriculture of the People’s Republic of China (GJFP201701505; GJFP2019019).
Author information
Authors and Affiliations
Contributions
YZ and ZX conceived and designed research. YZ and LD conducted experiments. ZY, DZ, JJ, and JQ contributed new reagents or analytical tools. YZ and LD analyzed data and wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, Y., Ding, L., Yan, Z. et al. Identification and Characterization of a Novel Carboxylesterase Belonging to Family VIII with Promiscuous Acyltransferase Activity Toward Cyanidin-3-O-Glucoside from a Soil Metagenomic Library. Appl Biochem Biotechnol 195, 2432–2450 (2023). https://doi.org/10.1007/s12010-021-03614-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03614-9