Published online Jun 26, 2021. doi: 10.4252/wjsc.v13.i6.521
Peer-review started: February 16, 2021
First decision: March 17, 2021
Revised: March 22, 2021
Accepted: May 20, 2021
Article in press: May 20, 2021
Published online: June 26, 2021
Stem cells are undifferentiated cells that can self-renew and differentiate into diverse types of mature and functional cells while maintaining their original identity. This profound potential of stem cells has been thoroughly investigated for its significance in regenerative medicine and has laid the foundation for cell-based therapies. Regenerative medicine is rapidly progressing in healthcare with the prospect of repair and restoration of specific organs or tissue injuries or chronic disease conditions where the body’s regenerative process is not sufficient to heal. In this review, the recent advances in stem cell-based therapies in regenerative medicine are discussed, emphasizing mesenchymal stem cell-based therapies as these cells have been extensively studied for clinical use. Recent applications of artificial intelligence algorithms in stem cell-based therapies, their limitation, and future prospects are highlighted.
Core Tip: This article reviews some important types of stem cells in clinical treatment including embryonic stem cells, induced pluripotent stem cells, induced tissue-specific stem cells, and adult stem cells. Furthermore, the article focuses on the clinical treatment of mesenchymal stem cells and the application of artificial intelligence in induced pluripotent stem cells, their limitations, and future prospects.
- Citation: Mukherjee S, Yadav G, Kumar R. Recent trends in stem cell-based therapies and applications of artificial intelligence in regenerative medicine. World J Stem Cells 2021; 13(6): 521-541
- URL: https://www.wjgnet.com/1948-0210/full/v13/i6/521.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i6.521
Stem cells hold great potential in regenerative medicine as these cells are endorsed with indefinite self-renewal characteristics and can be differentiated into any cells of the body. These cells express specific markers and are karyotypically normal. Based on the differentiation potential, stem cells can be categorized as totipotent, pluripotent, multipotent, or unipotent[1,2]. Totipotent cells are those that can form the entire organism. In animals, only the zygote is totipotent, and therefore, cannot be used for therapeutic purposes. The pluripotent embryonic stem cells (ESCs) are isolated from the inner cell mass (ICM) of the embryo's blastocyst. These pluripotent cells can differentiate into all cells, including germ cells but not the entire organism. The multipotent cells can generate a definite group of cells, e.g., hematopoietic stem cells (HSCs) or mesenchymal stem cells (MSCs). The unipotent stem cells have restricted potential and can differentiate into a single cell type, e.g., a neuronal stem cell. Stem cells have been classified according to the origin into ESCs and tissue-derived stem cells[3,4]. The tissue-derived stem cells may be adult stem cells (ASCs) isolated either from bone marrow, peripheral blood[5], adipose tissue[6], dental pulp[7], skeletal muscle[8], skin[9], neural tissue[10], liver[11], heart[12], pancreas[13] or intestine[14]. ASCs have reduced potency compared to ESCs. They are still preferred over ESCs for regenerative medicine because of ethical issues associated with ESCs. The fetal stem cells can be isolated from either fetal tissue like blood, spleen, liver, kidney, or extraembryonic sources like an amnion, amniotic fluid, umbilical cord, Wharton's jelly, and placenta. The HSCs and MSCs isolated from fetal tissues or extraembryonic sources have greater potential than their adult counterparts. Therefore, these cells are considered a "half-way house" between the ESCs and ASCs in terms of plasticity[15,16]. Induced pluripotent stem cells (iPSCs) are produced by reprogramming differentiated cells into an undifferentiated state[17]. These reprogrammed stem cells are artificially produced cells with properties of ESCs. Induced tissue-specific stem (iTS) cells have recently been produced by incomplete reprogramming and tissue-specific selection[18,19].
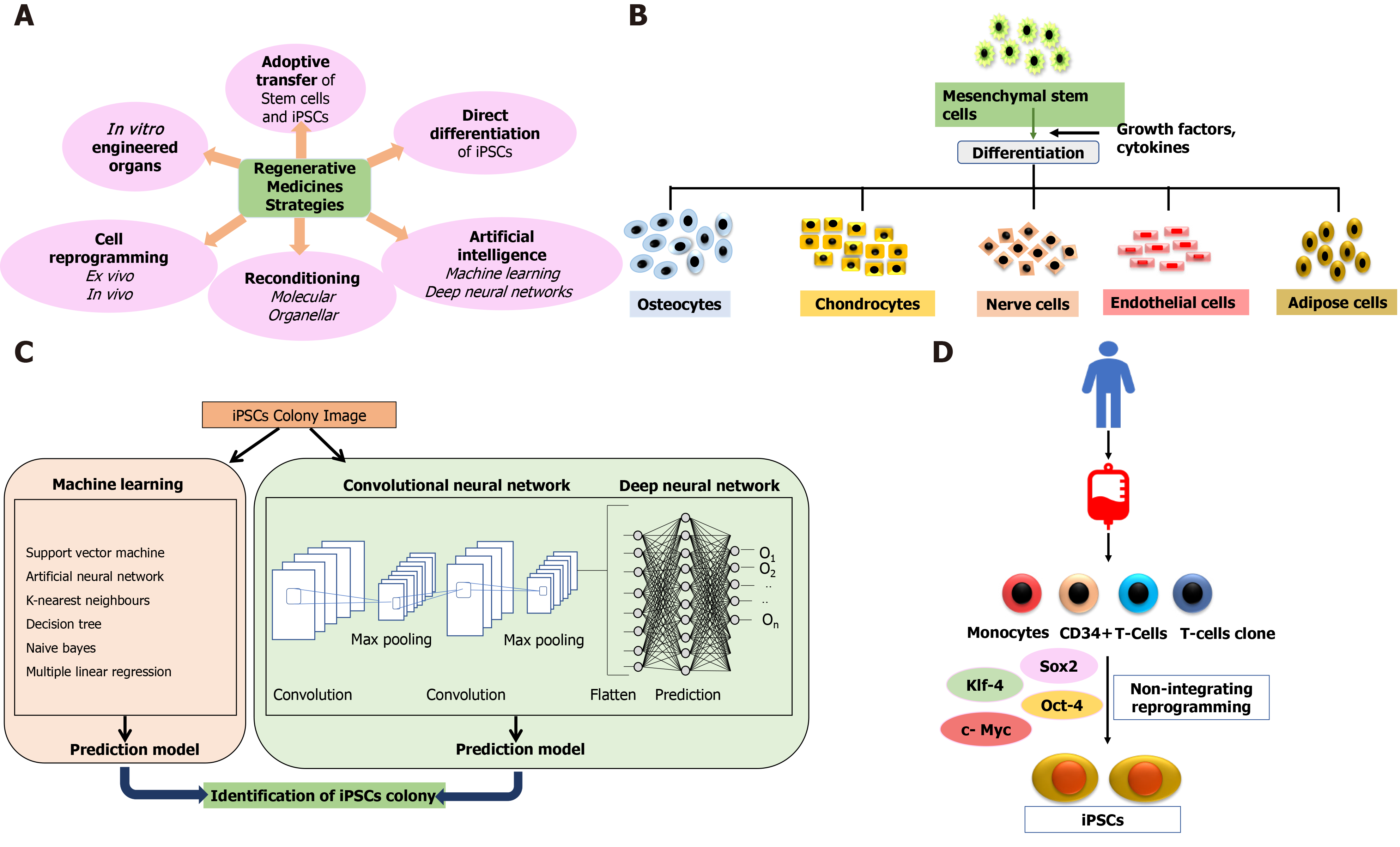
Cell therapy applies the biochemical and biophysical properties of stem cells to generate healthy tissues and repair damaged organs, preventing them from further damage[20]. The bone marrow serves as a source of stem cells from where HSCs, neural stem cells, and MSCs can be derived for therapeutic purposes[21]. These cells (termed adult cells) are non-pluripotent, limiting them to proliferate in only those types of tissues from which they have been isolated. Further advancement in stem cell therapy happened with identifying pluripotency of ESCs in 1998[22]. As the generation of ESCs involves the exploitation of embryos, this raised major ethical issues and limited the scope of stem cell-based therapies yet again until the creation of iPSCs in 2007[23-25]. Stem cell therapy holds the solid potential for combat transplantation-related issues such as graft rejection or tissue insufficiency and opens the door for precision medicines. Figure 1 depicts regenerative strategies, MSC differentiation, iPSCs, and the role of artificial intelligence (AI) in regenerative medicines.
The non-pluripotent nature of available ASCs has restrained the scope of cell therapy in regenerative medicine. The ESCs have their share of ethical and fewer availability issues. These limitations called for an advancement in cell therapy and led to the creation of iPSCs. The genetic reprogramming of ASCs has imparted them with ESC-like functional similarity and pluripotency, generating iPSCs[26,27]. iPSCs have gained popularity in multiple facets of cell-based therapies by serving as an unlimited source of any cell type of interest[28]. It has enabled the employment of iPSCs as novel human disease models[29,30], which have been applied to drug discovery as well as the fields of precision medicine and regenerative medicine[31]. Several preclinical studies in animal models have established MSC suitability for regenerative medicine[32]. Although several clinical trials have been carried out with stem cells to treat genetic disorders, autoimmune diseases, or degenerative disorders, several challenges and limitations are encountered in human clinical translation, which are being addressed to improve the regenerative potential of these cells.
ESCs exhibit pluripotency and differentiate into the three germ layers: ectoderm, mesoderm, and endoderm. These cells are derived from the ICM layer of the embryo's blastocyst. Mouse ESC was first derived in 1981 by Evans and Kaufman[33] in the United Kingdom and Martin[34] in the United States from the ICM of the blastocyst (d2.5). Human ESCs (hESCs) were derived by Thomson and colleagues isolated from preimplantation blastocysts[22]. hESCs have been an excellent source of pluripotent cells for therapeutic use[35]. Derivation of pluripotent ESCs from the blastocyst’s ICM layer is usually done by a standardized immunosurgery technique[36]. The ESCs are isolated and seeded on feeder layers in culture plates. The cells may be characterized for pluripotency markers by immunostaining using specific antibodies to octamer-binding transcription factor ¾ (Oct3/4), stage-specific embryonic antigen 3 (SSEA-3), SSEA-4, TRA-1-60, and TRA-1-81 or by assessing alkaline phosphatase activity. ESCs are karyotypically normal and possess a high telomerase activity[37,38]. However, there are severe concerns in using ESC in regenerative medicine despite being a promising candidate. Severe ethical concerns prevail in using human embryos for the isolation of hESCs[39]. Different legal guidelines are governing ESC research in various countries. In the United States, the destruction of human embryos for any form of research is banned. According to the guidelines, hESC-lines derived before August 9, 2001 can be used for research. The development of hESC therapies is restricted, and most research studies are focused on animals[40]. In the United Kingdom, research using hESCs derived from discarded embryos in in vitro fertilization clinics is allowed; however, hESC research is prohibited in Italy[41-43].
It is also essential to analyze the safety issues associated with ESCs in regenerative medicine. The ESCs can differentiate into any cell type of the body. However, when these undifferentiated cells are implanted in vivo, this plasticity poses the risk of developing teratomas and tumors[44-47]. The alternative method is to differentiate the undifferentiated ESCs in vitro into a specific cell type along a lineage and then transplant the differentiated cell in vivo. hESC-derived cardiomyocytes, when transplanted in mice, did not result in teratoma[48]. However, in certain instances, the transplanted progenitor cells continue to proliferate, as, for example, nestin+ dopaminergic neurons derived from hESCs continue to proliferate in the striatum[49]. The screening of the undifferentiated cells by specific markers and their subsequent purification before transplantation may solve the problem.
There are some reported methods to overcome the risk of tumorigenesis, such that ESCs can be used for regenerative therapies[50]. One of the recent findings involves cluster of differentiation 133 (CD133) (prominin 1), a transmembrane protein generally expressed on cancer stem cells is highly expressed on hESCs. CD133-deficient knock-out hESC line retained the capacity to differentiate into the three embryonic germ layers in vivo. Still, the proliferating potential is reduced and results in reduced teratoma formation[51]. Therefore, CD133 may be used to sort ESCs for transplan
Despite the safety issues, hESC-derived progenitor cells are still considered promising candidates in regenerative medicine under controlled conditions[53]. The first approval of the hESC trial for spinal cord injury was received in 2009, in which hESC-derived oligodendrocytes progenitor cells were used[54]. The hESC-based clinical trials have been performed for the treatment of macular degeneration with some positive results in follow-up studies[55-58], diabetes mellitus[59], and ischemic heart disease[60]. One clinical trial has been approved for Type 1 diabetes produced by the company ViaCyte (ClinicalTrials.gov Identifier: NCT03163511, NCT02239354)[61]. These pancreatic progenitor cells are produced from human pluripotent stem cells in vitro and differentiate into beta cells after transplantation in an immune isolation device in vivo[62]. Clinical trials for Parkinson’s disease with hESCs are being conducted in Australia (NCT02452723) and China (NCT03119636)[63].
iPSCs were first successfully generated by Takahashi and Yamanaka[64] in 2006 by inserting the reprogramming factors known as “Yamanaka factors,” Oct4/3, Sox2, Klf4, and c-Myc in mouse fibroblast cells. Initially, Yamanaka and his group (Okita et al[65]) started transducing mouse fibroblasts with a recombinant retrovirus carrying 24 genes responsible for maintaining the ESC characteristics. The mouse fibroblasts were selected by antibiotic-resistant gene cassette under the promoter, Fbx15, which is active only in the ESCs. The number of genes was narrowed down to ten and finally to four genes, Oct3/4, c-Myc, Sox2, and Klf4. The first generation induced pluripotent cells selected by Fbx15 possessed unlimited self-renewal and differentiation capacity, produced embryoid bodies and fetal chimeras but failed to produce adult chimeras. However, the DNA methylation pattern, post-translational modifications, and epigenetic changes revealed that the generated iPSCs were intermediate between fibroblasts and ESCs. Yamanaka and his group further generated second-generation iPSCs using the selection for Nanog instead of Fbx15 selection. The second-generation iPSCs showed greater ES cell-like characteristics, DNA methylation pattern, and germline competence[65]. However, 20% of the chimeric mice developed cancer as two of the genes, c-Myc, and Klf4, are oncogenic. Human-induced pluripotent cells were generated from somatic cells in 2007 by two independent groups simultaneously by introducing Oct3/4 and Sox2 with either Klf4 and c-Myc or Nanog and Lin28[66]. The latter group has shown that reprogramming of human somatic cells is possible even when the reprogramming factors are not integrated into the genome. The use of non-integrated episomal vectors makes these cells more suitable for clinical use[67]. iPSCs have also been derived from peripheral blood mononuclear cells[68]. Adenovirus has also been used vector for the delivery of reprogramming factors.
However, virus-mediated delivery systems sometimes threaten iPSCs' clinical use due to insertional mutagenesis, mainly caused by cMyc[69-71]. RNA, proteins, and small molecules enhance iPSCs' efficiency and safety. Reprogramming of somatic cells by mRNA or microRNA became very successful and effective[72-75]. It has been reported that activation of the innate immune system enhances the efficiency of induced pluripotent cells by mRNA transfection[76]. Insertion of recombinant proteins such as Oct 4, sex-determining region Y-box 2 (Sox2), Kruppel-like factor 4 (Klf4), and cMyc can be used to reprogram somatic cells to induce pluripotent cells[77-79]. The protein-based approach has been used to generate dopaminergic neurons from iPSCs to treat Parkinson’s disease in rats[80].
The iPSCs have potential similar to that of ESCs, and additionally overcome the ethical concerns associated with ESC research and clinical use. The iPSCs have gained much attention in recent years because of their advancement in regenerative medicine, organoid formation, and scope for personalized therapies. The first human clinical trial with iPSCs was done for macular degeneration in which retinal pigmented epithelial cells generated from autologous iPSCs were transplanted into a patient[81]. There have been several in vitro[82,83] and in vivo preclinical studies investigating the safety and efficacy of iPSCs[84-87]. A human clinical trial was recently conducted to treat Parkinson’s disease in Kyoto, Japan, by transplanting dopaminergic neurons generated from iPSCs. The clinical-grade iPSCs are produced from cells taken from healthy volunteers[88]. Autologous iPSC-derived dopaminergic neurons transplanted in a patient with Parkinson’s disease retained the function until 2 years without any adverse effect[89]. Autologous iPSCs are more advantageous to avoid immunological rejection, but the development of autologous iPSC is time-consuming and costly.
The iPSC technology has enormous potential in regenerative medicine. However, more interventional studies[90] must be conducted to address the challenges of routine clinical applications of these cells e.g., genomic instability[91], carcinogenicity[92], immunological rejection[93]. Humanized mouse models, e.g., a mouse with human immune cells, may be developed in the future to investigate the immunogenicity of human pluripotent stem cells[94].
iTS are produced by incomplete reprogramming of somatic cells by transient overexpression of reprogramming factors by plasmids and performing a tissue-specific selection. These cells have the potential to self-renew but also express tissue-specific markers. iTS have been produced from mouse pancreatic cells, which can self-renewal and express pancreatic tissue-specific transcription factor, Pdx1[13]. These generated iTS can differentiate into insulin-producing cells more efficiently than ESCs or iPSCs and, probably, can be utilized to treat diabetes. iTS with neural stem cell-like characteristics have also been reported[95-98]. Teratoma formation is not reported when iTS are transplanted in nude mice. In this respect, iTS are advantageous over the ESCs or iPSCs in terms of clinical applications due to the risk of tumorigenicity associated with the use of pluripotent stem cells. These cells are “incompletely reprogrammed” cells and have different methylation patterns than ESCs or iPSCs. The iTS retain the donor tissue’s epigenetic memory and can be explored as a potential candidate for cell replacement therapy.
Fetal stem cells are collected from aborted fetal tissues and extraembryonic structures like amniotic fluid, umbilical cord, Wharton’s jelly, and placenta[99]. The fetal stem cells are multipotent HSCs or MSCs. HSCs collected from fetal bone marrow or umbilical cord express CD34 and CD45 Like adult HSCs but show greater proliferating capacity, low immunogenicity, and lower risk of graft vs host disease (GvHD) compared to adult HSCs. Fetal MSCs can be isolated from fetal blood, bone marrow, liver, lung, or pancreas. These cells have more differentiation capacity than the adult MSCs. Fetal MSCs have active telomerase and express low levels of human leukocyte antigen (HLA) I and lack intracellular HLA II[100]. First trimester fetal MSCs express baseline levels of pluripotent stem cell markers such as Oct4, Nanog, Rex1 SSEA3, SSEA4, Tra-1-60, and Tra-1-81. Umbilical cord blood MSCs are easy to harvest and can be stored under controlled conditions for longer periods for future clinical use[101]. Although the ethical concerns associated with fetal stem cells are minimal, there are several reports on the adverse effects of fetal stem cells in vitro in animals and humans[102]. Thromboembolism has been reported in patients who received transplantation of umbilical cord blood MSCs[103]. Placental-derived MSCs express higher levels of tissue factor[104,105], which aggravates the thrombotic events in patients infused with these MSCs for treatment of Crohn’s disease[106]. The ASCs are less prevalent and undifferentiated cells present in various adult tissues with a primary role of repair and maintenance of residing tissues.
ASCs are derived from different tissues and have limited potency compared to the ESCs or iPSCs. ASCs are named depending on the tissue of origin, e.g., HSCs, pancreatic stem cells, corneal stem cells, etc. HSCs were the first multipotent ASCs isolated from the bone marrow[107]. HSC transplantation is used as therapy for several malignant and non-malignant disorders and autoimmune diseases. These cells are also used for the recovery of patients undergoing chemotherapy and radiotherapy[108]. The allogeneic transplantation requires matching HLA, Class I, and Class II between donor and recipient[109]; however, there are risks of GvHD[110]. The emergence of more modern and less toxic methods of treatments replaces HSC transplantation in hematologic malignancies. Recent reports suggest that risks of bloodstream infections caused by Gram-negative bacteria are associated with allogeneic hematopoietic transplantation[111,112]. Hemorrhagic cystitis is another complication that has been reported in patients post-HSC transplantation[113].
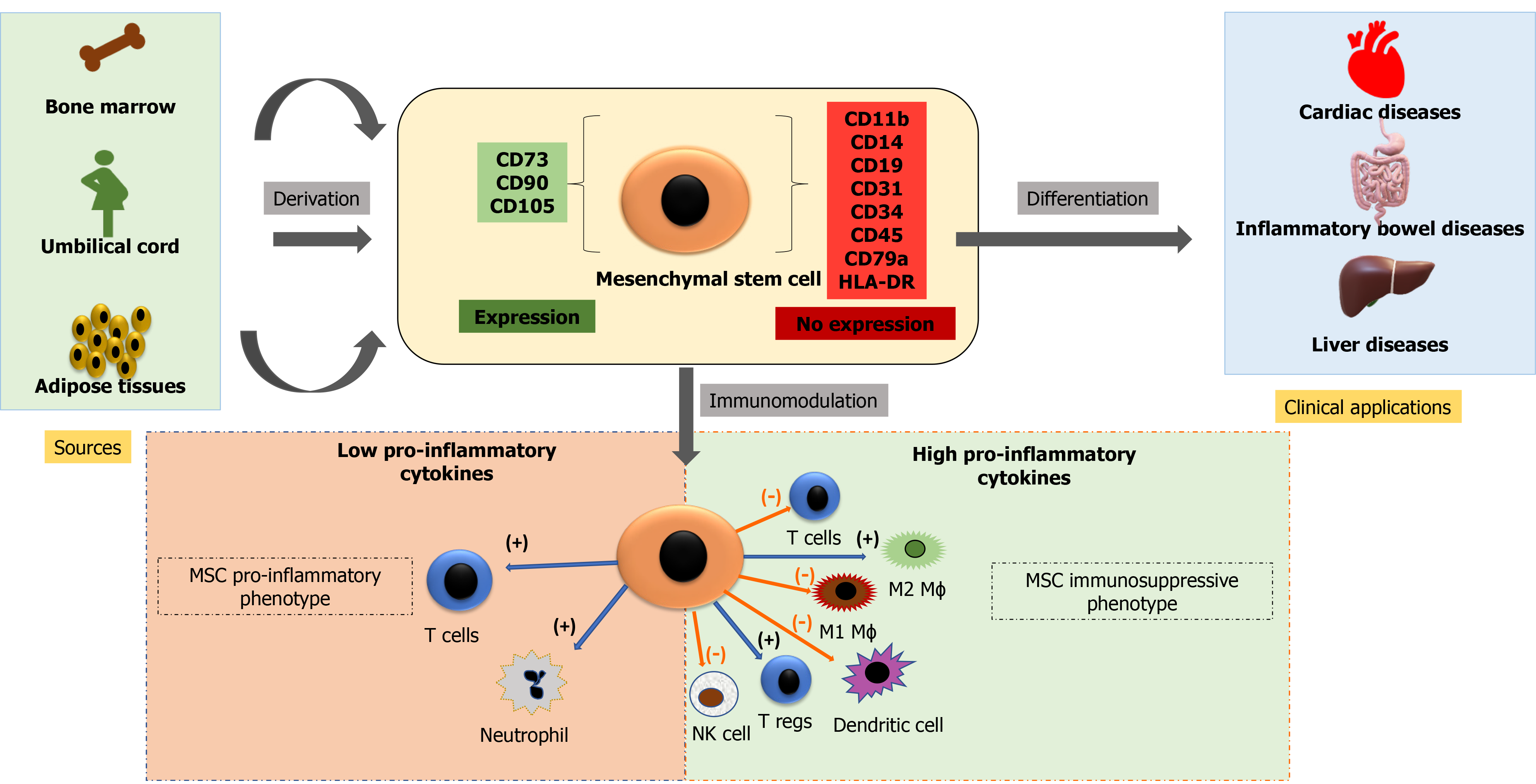
Cell-based therapy using MSCs is currently an essential domain of research. MSCs may be isolated from various sources, including bone marrow, adipose tissues, dental pulp, peripheral blood, synovium, and extraembryonic sources, as described earlier. These cells are plastic adherent and multipotential and can differentiate into bones, cartilage, fat tissues, muscles, and tendons[114]. The International Society for Cellular Therapy defined MSCs as plastic adherent cells expressing CD73, CD105, CD90 (≥ 90%), and not expressing hematopoietic markers CD34, CD45, CD14, CD19, and HLA-DR (≤ 2%) and have the potential of multilineage differentiation to osteogenic, adipogenic and chondrogenic lineage[115]. There is, however, no single marker to identify MSCs from various sources[116-118]. Figure 2 shows immune-modulatory characteristics and the ability of differentiation of MSCs.
The three primary characteristics of MSCs are: Differentiation into specific cell types and their incorporation into tissues that make it suitable for regenerative medicine; secretion of cytokines and exosomes that stimulate cell growth and proliferation and modulate inflammation; and direct contact with the host tissue and regulate effector function[119,120]. MSCs are therapeutically more successful due to the multipotentiality, immunomodulatory, anti-inflammatory, efficient homing capacity to injured sites, and minimum ethical issues[121-123]. However, there are disparities in MSC potency and pharmacological functionality, depending on tissue sources, cell handling, method of harvest, cultural expansion, dose, and route of delivery[124].
The most important sources of MSCs for clinical trials are bone marrow and adipose tissues[125]. Although MSCs isolated from the bone marrow have been extensively studied. However, there are several challenges in the clinical use of these cells. Cells collected from bone marrow are contaminated with HSCs, and a very small fraction (0.001%-0.01%) of MSCs are harvested from bone marrow. The cells isolated also show early signs of senescence during culture[126]. Although the collection of bone marrow aspirate is considered safe, certain complications and morbidity have been reported during collection from the sternum and posterior iliac crest[127]. Adipose tissue-derived stem cells are another potential source of MSCs by less invasive procedures[128]. Adipose tissue is abundant in the body and can be easily collected by liposuction, and the yield of stem cells is comparatively greater without any adverse effects. Adipose tissue-derived stem cells have more stability in the culture and have greater differentiation potential to osteocytes, chondrocytes, adipocytes, cardiomyocytes, and neurocytes[129]. Several studies have reported adipose tissue-derived MCSs' efficacy and safety for regenerative medicine[130,131]. The other sources include dental pulp, tendon, or from the perivascular fraction of any tissue[132].
MSCs have immunomodulatory properties and can reduce the inflammatory response. These cells can modulate the function of the innate and adaptive immune response. MSCs have very low levels of major histocompatibility complex and reduced expression of FasL (Fas Ligand) or costimulatory signals like B7, CD40, or CD40L[133]. MSCs secrete extracellular vesicles containing various growth factors and cytokines that suppress B lymphocyte and T lymphocyte function and maturation of dendritic cells while activating T regulatory cells. MSCs also secrete angiogenic, antiapoptotic, and antioxidative effects[134]. Table 1 summarizes the discovery time, sources, advantages, disadvantages, current clinical applications and prospects of various stem cells.
| Type of stem cell | Discovery time | Source | Advantages | Disadvantages | Clinical applications and prospects |
| Embryonic stem cells | mESC was first derived in 1980 by Evans and Kaufman[33] in the United Kingdom and Martin[34] in the United States. hESC was derived by Thomson et al[22] isolated from preimplantation blastocysts in 1998 | ICM of embryo | Maximum potency and these cells have the potential to differentiate into any cell type of the body | Ethical concerns, risk of developing teratomas and tumors when these undifferentiated cells are implanted in vivo[44-47] | Spinal cord injury[54], macular degeneration[55-58], diabetes mellitus[59], ischemic heart disease[60] |
| Induced pluripotent stem cells | Induced pluripotent stem cells were first successfully generated by Takahashi and Yamanaka[64] in 2006 | Fibroblast cells | These cells have the potential to differentiate into any cell type of the body. Overcomes the ethical concerns associated with embryonic stem cell research and clinical use. Organoid formation, and scope for personalized therapies | Genomic instability, carcinogenicity, immunological rejection | Macular degeneration[81] and Parkinson's disease[89] |
| Fetal stem cells | First isolated and cultured by John Gearhart and his team at the Johns Hopkins University School of Medicine in 1998[185] | Umbilical cord blood cells | High availability and reduced ethical concerns. Higher expansion rate. Possess osteogenic differentiation capabilities. Produce 2.5-fold more insulin than bone marrow derived cells | May not have adipogenic potential | Pancreatic islet cell generation in vitro. GvHD and systemic lupus erythematosus |
| Amniotic fluid and placenta | Harvested with minimal invasiveness | No clinical trials have yet been conducted to assess the safety and effectiveness of these stem cells | Potential treatment for nerve injuries or neuronal degenerative diseases. Bladder regeneration, kidney, lung, heart, heart valve, diaphragm, bone, cartilage and blood vessel formation. Treatment for skin and ocular diseases, inflammatory bowel disease, lung injuries, cartilage defects, Duchenne muscular dystrophy, and stroke. Also used in peripheral nerve regeneration | ||
| Adult stem cells | |||||
| Hematopoietic stem cells | First discovered for clinical use in mice in 1950’s and for clinical use in human in 1970[186,187] | Bone marrow | Multipotent cells | Risks of GvHD[110]. Risks of bloodstream infections caused by Gram-negative bacteria associated with allogeneic hematopoietic transplantation[111,112]. Hemorrhagic cystitis is another complication that has been reported in patients post hematopoietic stem cell transplantation[113] | Hematopoietic stem cell transplantation is used as therapy for several malignant and non-malignant disorders and autoimmune diseases. These cells are also used for the recovery of patients undergoing chemotherapy and radiotherapy[108] |
| Mesenchymal stem cells | First derived in 1970 and first report of clinical use in 2004[188] | Bone marrow | Potential to differentiate osteocytes, chondrocytes, adipocyte. Multipotentiality, immunomodulatory, anti-inflammatory, efficient homing capacity to injured sites, and minimum ethical issues[121-123] | Procurement of cells from this source is often painful and carries the risk of infection. Cell yield and differentiation potential is dependent on donor characteristics | Generation of pancreatic cells in vitro. Orthopedic conditions characterized by large bone defects, including articular cartilage repair and osteoarthritis, rheumatoid arthritis. BM-MSCs may also be used to treat non-unions, osteonecrosis of the femoral head and to promote growth in osteogenesis imperfecta. Potentially promising treatment for myocardial infarction, GvHD, systemic lupus erythematosus and multiple sclerosis |
| First derived in 2001[185] | Adipose tissue isolated from liposuction, lipoplasty or lipectomy materials | This source results in the isolation of up to 500 times more stem cells than BM (5 × 103 cells from 1 g of AT). AT is accessible and abundant and secretes several angiogenic and antiapoptotic cytokines. The immunosuppressive effects of AT-MSCs are stronger than those of BM-MSCs | Cells from this source have inferior osteogenic and chondrogenic potential in comparison to BM-MSCs | Immunosuppressive GvHD therapy. Potential for cell-based therapy for radiculopathy, myocardial infarction, and neuropathic pain. Cosmetic/dermatological applications. Successfully used in the treatment of skeletal muscle-injuries, meniscus damage and tendon, rotator cuff and peripheral nerve regeneration | |
MSCs have been utilized for preclinical and clinical studies for a wide range of diseases owing to multipotentiality, immunomodulation, and regeneration. The first clinical trials involved the infusion of MSCs post high dose chemotherapy and reversing GvHD, which is resistant to steroids[135]. To date, numerous clinical trials have been undertaken using MSCs for various diseases like myocardial infarction, Crohn’s disease, multiple sclerosis, diabetes, GvHD, amyotrophic lateral sclerosis, arthritis, neurodegenerative disorders, trauma, coronavirus disease 2019, and many more[136] (Table 2). According to recent reports, 10000 patients have undergone treatment with MSCs for different diseases, and 1094 clinical trials are registered at present in different phases[137]. Due to the relative ease of isolation and efficacy, the most prevalent source of MSCs for therapeutic purposes remains bone marrow, followed by the umbilical cord and then adipose tissue. Placental MSCs represent less than 2% of all clinical trials, and then MSCs from other sources are utilized[138].
| Disease category | Target disease | Clinical trial phase | Cell source | Company | Product name | ID No. | Status |
| GvHD | GvHD | Phase III | Mesenchymal stem cells (allogenic bone marrow derived) | Osiris Therapeutics | Prochymal | NCT00366145 | Approved via Notice of Compliance with conditions (NOC/c)[32] |
| Pediatric (GvHD, Grade III and IV) | Phase III | Mesenchymal stem cells (allogenic bone marrow derived) | Mesoblast | Remestemcel-L (Ryoncil™) | NCT02336230 | Prescription Drug User Fee Act (PDUFA) set by US FDA action and Remestemcel-L will be commercially available in the United States (if approved)[124,139-141] | |
| Crohn’s disease | Phase III | Autologous AT-MSC | Cellerix | - | NCT00475410 | Completed in 2009 but failed | |
| Phase III | Allogenic, AT-MSC | TiGenix | Alofisel® | NCT01541579 | Approved in 2018, by the European Medicines Agency[142, 143] | ||
| Cardiovascular diseases | Chronic advanced ischemic heart failure | Phase III | Autologous BM-MSC | - | - | NCT01768702 | Beneficial but not approved yet, further studies need to be undertaken[144-146] |
| Autoimmune diseases | Systemic lupus erythematosus | Phase I/II | Allogenic BM-MSC, UC-MSC | - | - | NCT01741857, NCT00698191 | Ongoing[147,148] |
| Type I diabetes | Phase I/II | Allogenic, UC-MSC combined with aulogous BM-MSC | - | - | NCT01374854 | Ongoing[149] | |
| Neurodegenerative diseases | Parkinson’s disease | Phase I/II | Allogenic BM-MSC | - | - | NCT02611167 | Completed but more interventional studies underway[150] |
| Alzheimer’s disease | Phase I | Allogenic UC MSC, Longeveron MSC, BM MSC | - | - | NCT04040348, NCT02600130, NCT02600130 | Ongoing[151] | |
| SARS-CoV-2 | COVID-19 | Phase II/III | BM-MSC, AT-MSC, Placenta derived MSC | Mesoblast, Athersys; Tigenix/Takeda; Pluristem | MultiStem; SPECELL | Ongoing[136,152] |
The conventional approach of regenerative medicine and cell-based therapies faced multiple challenges related to the enormous amount of data analysis, rising quantity and complexity, standardization methods, manual errors, and extremely difficult data handling. This further complicates the process of conclusion derivation and decision-making practices without the risk of errors involved. The implementation of iPSCs in different experimental and therapeutic approaches requires identifying iPSC-derived cells, a thorough estimation of iPSC quality, and characterization of the cell type. Evaluation of colony morphology through a manual approach is a tedious and error-prone process, and it is not feasible for large-scale cultures. Various study groups have implemented different AI algorithms to install an automated approach for accurate segmentation and colony quality estimation to overcome this limitation.
AI raised major hopes in this aspect and became a synergistic approach that augments human expertise[153-155]. AI refers to an imitation of human intelligence that imparts to a machine the ability to interpret and learn from external data[156] and later use those learnings to anticipate and do self-amendment in similar or novel scenarios[157]. AI uses automated algorithms to solve problems and assist in tasks like data mining and analyzing enormous amounts of datasets, observe patterns and predict outcomes that would not be possible by human intelligence[158]. AI-based technologies potentially aid clinical decision support in real-time, resulting in improved precision medicine[159].
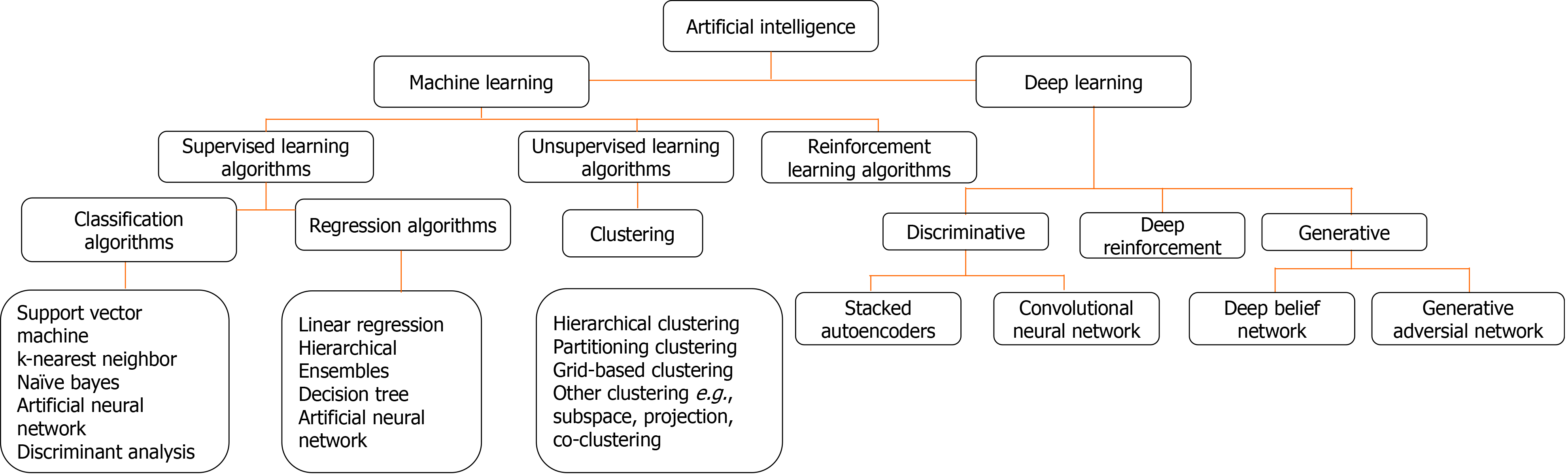
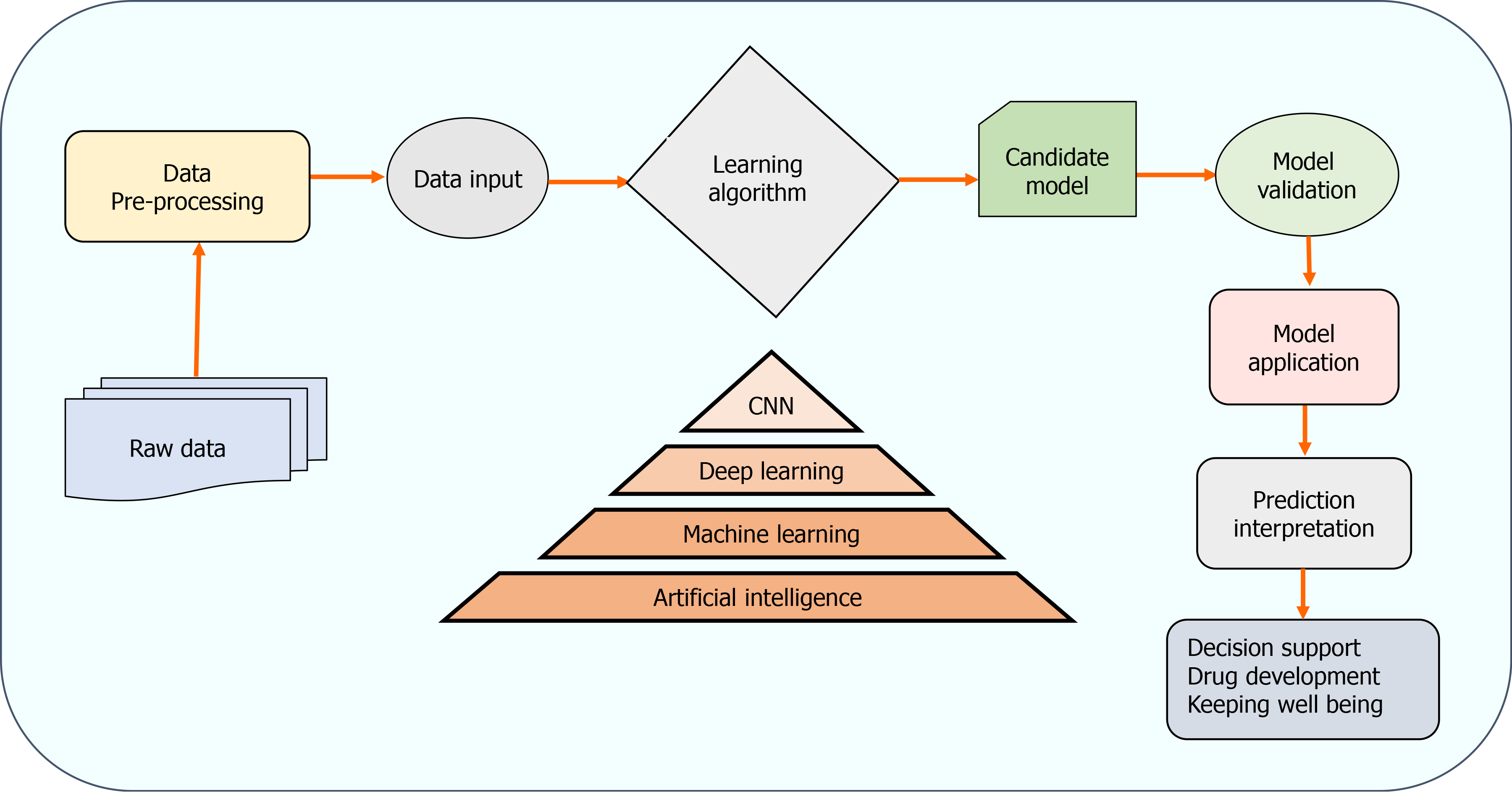
The two AI algorithms which are most widely adopted in the healthcare industry are machine learning (ML) and deep learning (DL) (Figure 3)[160,161]. ML refers to a data-based algorithm that allows the software to learn from first-hand information and become "intelligent" enough to perform predictive analysis and classification without being programmed for it[162]. The ML learning methods have been categorized into three types: supervised, unsupervised, and reinforcement learning. Supervised learning holds most of the ML tasks, and its training dataset consists of class labels. The label is a specific known outcome of interest, and the ML algorithm finds the best way to predict that outcome[163]. Unsupervised learning predicts the outcome without any prespecified labels and predicts the unknown patterns within the data. The supervised ML algorithms used in the field of medicine commonly include artificial neural network (ANN), support vector machine (SVM), naïve bayes, random forest (RF), k-Nearest Neighbors, decision tree (DT), and adaptive boosting (Adaboost), etc.[164]. Among these, ANN is the most widely used ML algorithm alongside SVM. ANN is a simulation of the human neuronal structure where neurons serve as the basic unit for communication and are arranged in sequential layers, with varying strength connections between layers. The input layer receives the training signal in pixel series or a speech series, relayed through the hidden layer where the data is analyzed. The concepts within the data are extracted to make the predictions and passed on to the output layer. The output layer further refines the data by performing classification/regression tasks[165]. DL's revolutionary concept emerged with an increased number of hidden layers, making it a more complex and, subsequently, even more, advanced concept of AI. DL techniques utilize these massive hidden neuronal units to automatically learn the complex relationship among the raw and noisy data, eliminating the tedious manual feature extraction required in ML algorithms[166]. Convolutional neural network (CNN) is a DL network robustly implemented in medical-imaging to perform image-classification and disease diagnosis tasks[167]. Figure 4 illustrates the general schema for AI-based prediction model development.
Joutsijoki et al[168] conducted a study focusing on an automated approach to identify iPSC colony images with 1608 × 1208 resolution by using ML classifiers- SVMs and k-NN. The authors also used scale-invariant feature transform descriptors in feature extraction. The k-NN classifier with Euclidean measure and equal weighting yielded the best result with an accuracy of 62.4%, which was a measured value compared to previous studies.
Kavitha et al[169] conducted a study to evaluate different automated texture features extracted from the segmented colony sections of iPSCs and use ML techniques to confirm their potential for characterization of colonies. They quantified 151 features obtained from the iPSCs images from phase-contrast microscope using moment-based, shape-based, spectral texture feature and statistical groups. A forward stepwise regression model was used to select the most relevant features for categorizing the colonies. Five ML classifiers- SVM, RF, MLP, Adaboost, and DT were used with 10-fold cross-validation to estimate the texture features within each texture-feature group and fused-feature groups to characterize diseased and healthy iPSC colonies. Based on one of their findings, SVM, RF, and Adaboost classifiers were concluded to exhibit superior performances compared to MLP and DT.
Several studies have reported the implementation of the DL algorithm through CNNs in iPSC studies. Kavitha et al[170], in their study, developed a vector-based CNN (V-CNN) using extracted features of the iPSC colony for finding out colony characteristics. They compared the V-CNN model with an SVM classifier using textural, morphological, and combined features. The study applied 5-fold cross-validation to examine the V-CNN performance. The precision, recall, and F-measure values were comparatively much higher than SVM (87%-93%). The V-CNN model was also subjected to determine the colony quality where the accuracy values on textural (91.0%), morphological (95.5%), and combined features (93.2%) bases were also found to be much higher than those of SVM values, which were 83.3%, 87.6%, and 83.4% respectively. In another study by Kusumoto et al[171], CNN's were utilized to identify iPSC-derived endothelial cells without immunostaining or lineage tracing. They obtained a dataset of 200 images from four experimental setups, of which 64 were applied for training alongside 160 for testing purposes. iPSC-derived endothelial cells' morphological descriptors (Phase-contrast images-based) were used to train the network. Its prediction validation was done by comparing with immunofluorescence staining for CD31, which is an endothelial cells marker. The method parameters were iteratively and automatically improved to obtain an error-free prediction. It was found that prediction accuracy was a function of the pixel size of the images and network depth in question. The k-fold cross validation also suggested that morphological features alone could be enough for optimizing CNNs, and they can deliver a high-value prediction. The next year, Waisman et al[172] used CNNs to separate pluripotent cells from initial differentiating cells. The authors used light microscopic images of PSCs to train the CNN model. Images of mouse-embryonic cells induced to epiblast-like cells were taken at different intervals after induction. The findings suggested that CNN can be trained to distinguish among differentiated and undifferentiated cells with an accuracy of 99%.
The well-established role of iPSCs in disease modeling and AI in therapeutics has also been exploited in several studies. Juhola et al[173] used iPSC-derived cardiomyocytes to study drug effects and their calcium transient signals with ML. Six iPSC-lines containing various catecholaminergic polymorphic ventricular tachycardia causing mutations were used to assess the drug-effect. The drug being studied was dantrolene after adrenaline stimulation by ML analysis of Ca2+ signals.
They identified the beats of transient signals with a previously proposed analytical algorithm[174], which recognizes signal abnormality depending upon whether the assessed cell signal has at least one abnormal transient peak-based on characteristics of a single peak. They computed 12 peak-variables for all identified signal-peaks. These data were used to classify the signals into various classes that correspond to those influenced by dantrolene or adrenaline. The algorithm's best classification accuracy was found to be nearly 79% suggesting a significant role of ML in the analysis of iPSC-cardiomyocytes drug effects.
In a similar study, Hwang et al[175] employed advanced ML techniques with an Analytical Algorithm to build an analytical pipeline for automatic evaluation of Ca2+ transient anomaly in cardiomyocytes. The pipeline was made up of peak detection, peak and signal abnormality assessment, and peak and signal variable detection. A peak-level SVM classifier was trained by using manual expertise. Two hundred cells were used as training data to train the SVM (cell-level), and other datasets of 54 cells were used to test the accuracy. The training and test accuracies were found to be 88% and 87%, respectively.
In a recent study, Nishino et al[176] developed a linear classification learning model to differentiate between iPSCs, ESCs, somatic cells, and embryonal carcinoma cells based on DNA methylation profiles. The accuracy of the ML model in identifying various cell types was found to be 94.23%. Furthermore, component analysis of the learned models identified the distinct epigenetic signatures of the iPSCs. Studies about recent AI-based stem cell therapies are summarized in Table 3.
| Study objectives | Applied AI algorithm | Important conclusions | Study group |
| iPSC-derived endothelial cells Identification without the application of molecular labelling using CNN | CNN | Prediction accuracy was a function of pixel size of the images and network depth. The k-fold cross validation suggested that morphological features alone could be enough for optimizing CNNs and they can deliver a high value prediction | Kusumoto and Yuasa[167] (2019) |
| Automated identification of the iPSC colony images quality | SVM, k-NN | k-NN yielded 62% of the accuracy which was found to be better than the previous studies of that time | Joutsijoki et al[168] (2016) |
| Assess automated texture descriptors of segmented colony regions of iPSCs and to check their potential | SVM, RF, MLP, Adaboost, DT | SVM, RF and Adaboost classifiers were concluded to exhibit superior classification ability than MLP and DT | Kavitha et al[169] (2018) |
| Develop a V-CNN model to distinguish the colony-characteristics on the basis of extracted descriptors of the iPSC colony | CNN | Recall, precision, and F-measure values by CNN were found to be comparatively much higher than the SVM. Colony quality accuracy was found to be 95.5% (morphological), 91.0% (textural) and 93.2% (textural) | Kavitha et al[170] (2017) |
| Use CNNs with transmitted light microscopy images to find out pluripotent stem cells from initial differentiating cells | CNN | CNN can be trained to distinguish among differentiated and undifferentiated cells with an accuracy of 99% | Waisman et al[172] (2019) |
| Use machine learning algorithms to analyze drug effects on iPSC cardiomyocytes | NB, KNN, LS-SVM, DT, multinomial logistic regression | Classification accuracy of the algorithm developed was found to be nearly 79% | Juhola et al[173] (2021) |
| To build an analytical procedure for automatic evaluation of Ca2+ transient abnormality, by applying SVM together with an analytical algorithm | SVM | The training and test accuracies were found to be 88% and 87% respectively | Hwang et al[175] (2020) |
| To develop a linear classification-learning model to differentiate among somatic cells, iPSCs, ESCs, and ECCs on the basis of their DNA methylation profiles | Jubatus (ML analytical platform) | The accuracy of the ML model in identifying various cell types was found to be 94.23%. Also, component analysis of the learned models identified the distinct epigenetic signatures of the iPSCs | Nishino et al[176] (2021) |
Apart from the above-mentioned studies that demonstrate applications of AI in cell culture stages, sufficient evidences are available that AI could play a significant role in predicting the MSC’s therapeutic outcomes[177-179]. Precise therapeutic outcome prediction of the MSC therapy could provide vital information for clinicians to assist them in decision support and decide the optimized treatment strategies. AI algorithms could be applied to optimize the clinical trials of innovative stem cell therapies for various diseases by precise treatment-planning for patients, clinical outcomes prediction, and patient recruitment, thereby reducing the complexity of the study and overall costs[180]. Machine and human intelligence together could have an exponentially high impact on the continual progress of stem cell-based therapy.
Regenerative medicines offer enormous potentials for better treatment of patients and quick recovery. However, there are certain drawbacks due to inefficient production, lengthy and complex processes, and human errors due to excessive human efforts. Understanding genetic components that influence the development of shape, size and orientation of an organ is extremely vital. Although the mechanism of most of the regenerative models is available with diagrams depicting the gene regulations, however, the stepwise dynamics to produce a particular shape of an organ are lacking. The AI-driven models and constructive algorithms could be a powerful solution for a deeper understanding of such mechanisms. These models could make the development of regenerative medicines automated and minimize human error factors.
The regenerative medicines' manufacturing has its own set of challenges, viz. efficient, cost-effective, large-scale production, lack of automation and quality control systems, and absence of closed and modular systems[181].
A vast number of datasets are published every year for experimental regenerative biology, but there is no international guideline for standard and high-quality datasets for regenerative medicines. Also, the tools for analyzing available datasets to get deeper insights and meaningful patterns are lacking. There have been some limited non-AI-based computational methods, platforms, and tools available to assist regenerative therapies. In an effort to derive such computational methods, Lobo and Levin developed a computational method to understand the physiological controls in planarian regeneration[182]. The exceptional ability of planaria to regenerate its body parts could be a potential model for leading regenerative medicine research. Another computational platform, KeyGene[183], can predict the tissue origin of various cell types. It could find out the equivalent stage of human PSC differentiation products along with the identification of stem cell derivatives. The KeyGene algorithm applies next-generation sequencing and microarray datasets and could be used to predict human adult tissue identity. This tool also helps monitor the cell-differentiation conditions and evaluate in-vitro cell-differentiation efficacy, thereby fetching improved protocol outcomes. CellNet[184] is among the recent computational tools that provide cell identity parameters, evaluate cell fate conversions, and rank suitable candidates for future interventions. However, it cannot differentiate between cell subtypes and cell heterogenicity, which remains a challenge to solve to date.
Nevertheless, AI-driven methods have emerged as an important component of stem cell research. Over the last decade, AI algorithms have advanced very rapidly, and along with enormous progress, methods to apply them have also enhanced subsequently. Many algorithms and tools can be expected in the recent future, which could efficiently assist stem cell-based regenerative medicines development, outcome prediction, and decision support to healthcare providers.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cao HC S-Editor: Gao CC L-Editor: Filipodia P-Editor: Xing YX
| 1. | Bacakova L, Zarubova J, Travnickova M, Musilkova J, Pajorova J, Slepicka P, Kasalkova NS, Svorcik V, Kolska Z, Motarjemi H, Molitor M. Stem cells: their source, potency and use in regenerative therapies with focus on adipose-derived stem cells - a review. Biotechnol Adv. 2018;36:1111-1126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 275] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 2. | Bissels U, Diener Y, Eckardt D, Bosio A. Characterization and classification of stem cells. In: Steinhoff G, editor. Regenerative Medicine – From Protocol to Patient. Cham, Switzerland: Springer International Publishing, 2016: 1-25. [Cited in This Article: ] |
| 3. | Ullah I, Subbarao RB, Rho GJ. Human mesenchymal stem cells - current trends and future prospective. Biosci Rep. 2015;35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 715] [Cited by in F6Publishing: 813] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 4. | Noguchi H, Saitoh I, Tsugata T, Kataoka H, Watanabe M, Noguchi Y. Induction of tissue-specific stem cells by reprogramming factors, and tissue-specific selection. Cell Death Differ. 2015;22:145-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 5. | Esposito MT. Blood factory: which stem cells? BMC Hematol. 2018;18:10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 6. | Zimmerlin L, Donnenberg VS, Rubin JP, Donnenberg AD. Mesenchymal markers on human adipose stem/progenitor cells. Cytometry A. 2013;83:134-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 173] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 7. | Stanko P, Altanerova U, Jakubechova J, Repiska V, Altaner C. Dental Mesenchymal Stem/Stromal Cells and Their Exosomes. Stem Cells Int. 2018;2018:8973613. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 8. | Hinken AC, Billin AN. Isolation of Skeletal Muscle Stem Cells for Phenotypic Screens for Modulators of Proliferation. Methods Mol Biol. 2018;1787:77-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Ojeh N, Pastar I, Tomic-Canic M, Stojadinovic O. Stem Cells in Skin Regeneration, Wound Healing, and Their Clinical Applications. Int J Mol Sci. 2015;16:25476-25501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 171] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 10. | Zhang J, Lu X, Feng G, Gu Z, Sun Y, Bao G, Xu G, Lu Y, Chen J, Xu L, Feng X, Cui Z. Chitosan scaffolds induce human dental pulp stem cells to neural differentiation: potential roles for spinal cord injury therapy. Cell Tissue Res. 2016;366:129-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 11. | Willemse J, Lieshout R, van der Laan LJW, Verstegen MMA. From organoids to organs: Bioengineering liver grafts from hepatic stem cells and matrix. Best Pract Res Clin Gastroenterol. 2017;31:151-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 12. | Klopsch C, Skorska A, Ludwig M, Gaebel R, Lemcke H, Kleiner G, Beyer M, Vollmar B, David R, Steinhoff G. Cardiac Mesenchymal Stem Cells Proliferate Early in the Ischemic Heart. Eur Surg Res. 2017;58:341-353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Saitoh I, Sato M, Soda M, Inada E, Iwase Y, Murakami T, Ohshima H, Hayasaki H, Noguchi H. Tissue-Specific Stem Cells Obtained by Reprogramming of Non-Obese Diabetic (NOD) Mouse-Derived Pancreatic Cells Confer Insulin Production in Response to Glucose. PLoS One. 2016;11:e0163580. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Barisas DAG, Stappenbeck TS. Intestinal Stem Cells Live Off the Fat of the Land. Cell Stem Cell. 2018;22:611-612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Abdulrazzak H, Moschidou D, Jones G, Guillot PV. Biological characteristics of stem cells from foetal, cord blood and extraembryonic tissues. J R Soc Interface. 2010;7 Suppl 6:S689-S706. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 16. | Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10:68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 414] [Cited by in F6Publishing: 631] [Article Influence: 126.2] [Reference Citation Analysis (0)] |
| 17. | Takahashi J. Strategies for bringing stem cell-derived dopamine neurons to the clinic: The Kyoto trial. Prog Brain Res. 2017;230:213-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Noguchi H, Miyagi-Shiohira C, Nakashima Y. Induced Tissue-Specific Stem Cells and Epigenetic Memory in Induced Pluripotent Stem Cells. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Miyagi-Shiohira C, Nakashima Y, Kobayashi N, Saitoh I, Watanabe M, Noguchi H. Characterization of induced tissue-specific stem cells from pancreas by a synthetic self-replicative RNA. Sci Rep. 2018;8:12341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Mahla RS. Stem Cells Applications in Regenerative Medicine and Disease Therapeutics. Int J Cell Biol. 2016;2016:6940283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 356] [Cited by in F6Publishing: 290] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 21. | Kumar R, Sharma A, Pattnaik AK, Varadwaj PK. Stem cells: An overview with respect to cardiovascular and renal disease. J Nat Sci Biol Med. 2010;1:43-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145-1147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11399] [Cited by in F6Publishing: 10061] [Article Influence: 387.0] [Reference Citation Analysis (0)] |
| 23. | Abad M, Mosteiro L, Pantoja C, Cañamero M, Rayon T, Ors I, Graña O, Megías D, Domínguez O, Martínez D, Manzanares M, Ortega S, Serrano M. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340-345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 349] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 24. | Volarevic V, Markovic BS, Gazdic M, Volarevic A, Jovicic N, Arsenijevic N, Armstrong L, Djonov V, Lako M, Stojkovic M. Ethical and Safety Issues of Stem Cell-Based Therapy. Int J Med Sci. 2018;15:36-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 359] [Cited by in F6Publishing: 402] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 25. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7589] [Cited by in F6Publishing: 6999] [Article Influence: 411.7] [Reference Citation Analysis (0)] |
| 26. | Glicksman MA. Induced Pluripotent Stem Cells: The Most Versatile Source for Stem Cell Therapy. Clin Ther. 2018;40:1060-1065. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 27. | Arjmand B, Goodarzi P, Mohamadi-Jahani F, Falahzadeh K, Larijani B. Personalized Regenerative Medicine. Acta Med Iran. 2017;55:144-149. [PubMed] [Cited in This Article: ] |
| 28. | Brodehl A, Ebbinghaus H, Deutsch MA, Gummert J, Gärtner A, Ratnavadivel S, Milting H. Human Induced Pluripotent Stem-Cell-Derived Cardiomyocytes as Models for Genetic Cardiomyopathies. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 29. | Nagoshi N, Okano H. Applications of induced pluripotent stem cell technologies in spinal cord injury. J Neurochem. 2017;141:848-860. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 30. | Yuasa S, Fukuda K. Recent advances in cardiovascular regenerative medicine: the induced pluripotent stem cell era. Expert Rev Cardiovasc Ther. 2008;6:803-810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Chen IY, Matsa E, Wu JC. Induced pluripotent stem cells: at the heart of cardiovascular precision medicine. Nat Rev Cardiol. 2016;13:333-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 130] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 32. | Galipeau J, Sensébé L. Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities. Cell Stem Cell. 2018;22:824-833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1009] [Cited by in F6Publishing: 1011] [Article Influence: 168.5] [Reference Citation Analysis (0)] |
| 33. | Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5956] [Cited by in F6Publishing: 5274] [Article Influence: 122.7] [Reference Citation Analysis (0)] |
| 34. | Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634-7638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3879] [Cited by in F6Publishing: 3466] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 35. | Levenberg S, Huang NF, Lavik E, Rogers AB, Itskovitz-Eldor J, Langer R. Differentiation of human embryonic stem cells on three-dimensional polymer scaffolds. Proc Natl Acad Sci USA. 2003;100:12741-12746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 532] [Cited by in F6Publishing: 441] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 36. | Solter D, Knowles BB. Immunosurgery of mouse blastocyst. Proc Natl Acad Sci USA. 1975;72:5099-5102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 410] [Cited by in F6Publishing: 384] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 37. | Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2078] [Cited by in F6Publishing: 1850] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 38. | Mukherjee S. Stem cell Characteristics and Applications. In: Singh R, Trivedi M. Biotechnology-Trends & Applications. Studium Press, LLC, USA, 2017: 277-299. [Cited in This Article: ] |
| 39. | De Trizio E, Brennan CS. The business of human embryonic stem cell research and an international analysis of relevant laws. J Biolaw Bus. 2004;7:14-22. [PubMed] [Cited in This Article: ] |
| 40. | Godfrey KJ, Mathew B, Bulman JC, Shah O, Clement S, Gallicano GI. Stem cell-based treatments for Type 1 diabetes mellitus: bone marrow, embryonic, hepatic, pancreatic and induced pluripotent stem cells. Diabet Med. 2012;29:14-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 41. | Stojkovic M, Lako M, Stojkovic P, Stewart R, Przyborski S, Armstrong L, Evans J, Herbert M, Hyslop L, Ahmad S, Murdoch A, Strachan T. Derivation of human embryonic stem cells from day-8 blastocysts recovered after three-step in vitro culture. Stem Cells. 2004;22:790-797. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 138] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 42. | Stojkovic M, Stojkovic P, Leary C, Hall VJ, Armstrong L, Herbert M, Nesbitt M, Lako M, Murdoch A. Derivation of a human blastocyst after heterologous nuclear transfer to donated oocytes. Reprod Biomed Online. 2005;11:226-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 128] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 43. | Heng BC, Tong GQ, Stojkovic M. The egg-sharing model for human therapeutic cloning research: managing donor selection criteria, the proportion of shared oocytes allocated to research, and amount of financial subsidy given to the donor. Med Hypotheses. 2006;66:1022-1024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 44. | Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, Agulnick AD, D'Amour KA, Carpenter MK, Baetge EE. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443-452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1331] [Cited by in F6Publishing: 1258] [Article Influence: 78.6] [Reference Citation Analysis (0)] |
| 45. | Prokhorova TA, Harkness LM, Frandsen U, Ditzel N, Schrøder HD, Burns JS, Kassem M. Teratoma formation by human embryonic stem cells is site dependent and enhanced by the presence of Matrigel. Stem Cells Dev. 2009;18:47-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 179] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 46. | Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11:268-277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 653] [Cited by in F6Publishing: 613] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 47. | Lee AS, Tang C, Rao MS, Weissman IL, Wu JC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19:998-1004. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 426] [Cited by in F6Publishing: 460] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 48. | Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015-1024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1672] [Cited by in F6Publishing: 1550] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 49. | Roy NS, Cleren C, Singh SK, Yang L, Beal MF, Goldman SA. Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat Med. 2006;12:1259-1268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 601] [Cited by in F6Publishing: 591] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 50. | Lee MO, Moon SH, Jeong HC, Yi JY, Lee TH, Shim SH, Rhee YH, Lee SH, Oh SJ, Lee MY, Han MJ, Cho YS, Chung HM, Kim KS, Cha HJ. Inhibition of pluripotent stem cell-derived teratoma formation by small molecules. Proc Natl Acad Sci USA. 2013;110:E3281-E3290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 177] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 51. | Kallas-Kivi A, Trei A, Maimets T. Lovastatin Decreases the Expression of CD133 and Influences the Differentiation Potential of Human Embryonic Stem Cells. Stem Cells Int. 2016;2016:1580701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 52. | Wang H, Gong P, Li J, Fu Y, Zhou Z, Liu L. Role of CD133 in human embryonic stem cell proliferation and teratoma formation. Stem Cell Res Ther. 2020;11:208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 53. | Song WK, Park KM, Kim HJ, Lee JH, Choi J, Chong SY, Shim SH, Del Priore LV, Lanza R. Treatment of macular degeneration using embryonic stem cell-derived retinal pigment epithelium: preliminary results in Asian patients. Stem Cell Reports. 2015;4:860-872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 248] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 54. | Alper J. Geron gets green light for human trial of ES cell-derived product. Nat Biotechnol. 2009;27:213-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 150] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 55. | Schwartz SD, Regillo CD, Lam BL, Eliott D, Rosenfeld PJ, Gregori NZ, Hubschman JP, Davis JL, Heilwell G, Spirn M, Maguire J, Gay R, Bateman J, Ostrick RM, Morris D, Vincent M, Anglade E, Del Priore LV, Lanza R. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt's macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet. 2015;385:509-516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 846] [Cited by in F6Publishing: 813] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 56. | Schwartz SD, Hubschman JP, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, Mickunas E, Gay R, Klimanskaya I, Lanza R. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713-720. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1016] [Cited by in F6Publishing: 927] [Article Influence: 77.3] [Reference Citation Analysis (0)] |
| 57. | Liu Y, Xu HW, Wang L, Li SY, Zhao CJ, Hao J, Li QY, Zhao TT, Wu W, Wang Y, Zhou Q, Qian C, Yin ZQ. Human embryonic stem cell-derived retinal pigment epithelium transplants as a potential treatment for wet age-related macular degeneration. Cell Discov. 2018;4:50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 58. | Vitillo L, Durance C, Hewitt Z, Moore H, Smith A, Vallier L. GMP-grade neural progenitor derivation and differentiation from clinical-grade human embryonic stem cells. Stem Cell Res Ther. 2020;11:406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 59. | Rezania A, Bruin JE, Xu J, Narayan K, Fox JK, O'Neil JJ, Kieffer TJ. Enrichment of human embryonic stem cell-derived NKX6.1-expressing pancreatic progenitor cells accelerates the maturation of insulin-secreting cells in vivo. Stem Cells. 2013;31:2432-2442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 196] [Cited by in F6Publishing: 193] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 60. | Menasché P, Vanneaux V, Hagège A, Bel A, Cholley B, Parouchev A, Cacciapuoti I, Al-Daccak R, Benhamouda N, Blons H, Agbulut O, Tosca L, Trouvin JH, Fabreguettes JR, Bellamy V, Charron D, Tartour E, Tachdjian G, Desnos M, Larghero J. Transplantation of Human Embryonic Stem Cell-Derived Cardiovascular Progenitors for Severe Ischemic Left Ventricular Dysfunction. J Am Coll Cardiol. 2018;71:429-438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 257] [Article Influence: 51.4] [Reference Citation Analysis (1)] |
| 61. | ViaCyte. A Safety, Tolerability, and Efficacy Study of VC-01™ Combination Product in Subjects With Type I Diabetes Mellitus. [accessed 2021 Jan 11]. In: ClinicalTrials.gov [Internet]. San Diego (CA): U.S. National Library of Medicine. Available from: https://clinicaltrials.gov/ct2/show/NCT02239354 ClinicalTrials.gov Identifier: NCT02239354. [Cited in This Article: ] |
| 62. | Memon B, Abdelalim EM. Stem Cell Therapy for Diabetes: Beta Cells vs Pancreatic Progenitors. Cells. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 63. | Barker RA, Parmar M, Studer L, Takahashi J. Human Trials of Stem Cell-Derived Dopamine Neurons for Parkinson's Disease: Dawn of a New Era. Cell Stem Cell. 2017;21:569-573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 221] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 64. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17989] [Cited by in F6Publishing: 17055] [Article Influence: 947.5] [Reference Citation Analysis (0)] |
| 65. | Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3332] [Cited by in F6Publishing: 3003] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 66. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14327] [Cited by in F6Publishing: 13579] [Article Influence: 848.7] [Reference Citation Analysis (0)] |
| 67. | Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1735] [Cited by in F6Publishing: 1628] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 68. | Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, Kim K, Miller JD, Ng K, Daley GQ. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476-5479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 447] [Cited by in F6Publishing: 413] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 69. | Okita K, Yamanaka S. Induced pluripotent stem cells: opportunities and challenges. Philos Trans R Soc Lond B Biol Sci. 2011;366:2198-2207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 177] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 70. | González F, Boué S, Izpisúa Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming à la carte. Nat Rev Genet. 2011;12:231-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 353] [Cited by in F6Publishing: 335] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 71. | Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678-684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 543] [Cited by in F6Publishing: 509] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 72. | Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618-630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1927] [Cited by in F6Publishing: 1831] [Article Influence: 130.8] [Reference Citation Analysis (0)] |
| 73. | Yakubov E, Rechavi G, Rozenblatt S, Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem Biophys Res Commun. 2010;394:189-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 155] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 74. | Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376-388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 900] [Cited by in F6Publishing: 869] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 75. | Mochiduki Y, Okita K. Methods for iPS cell generation for basic research and clinical applications. Biotechnol J. 2012;7:789-797. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 76. | Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547-558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 275] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 77. | Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472-476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1350] [Cited by in F6Publishing: 1202] [Article Influence: 80.1] [Reference Citation Analysis (0)] |
| 78. | Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Schöler HR, Duan L, Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381-384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1309] [Cited by in F6Publishing: 1208] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 79. | Liu X, Chen J, Firas J, Paynter JM, Nefzger CM, Polo JM. Generation of Mouse-Induced Pluripotent Stem Cells by Lentiviral Transduction. Methods Mol Biol. 2019;1940:63-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 80. | Rhee YH, Ko JY, Chang MY, Yi SH, Kim D, Kim CH, Shim JW, Jo AY, Kim BW, Lee H, Lee SH, Suh W, Park CH, Koh HC, Lee YS, Lanza R, Kim KS. Protein-based human iPS cells efficiently generate functional dopamine neurons and can treat a rat model of Parkinson disease. J Clin Invest. 2011;121:2326-2335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 170] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 81. | Okano T, Sawa Y, Barber E, Umezawa A. Regenerative therapy by fusion of medicine and engineering: First-in-human clinical trials with induced pluripotent stem cells and cell sheet technology: A report of the Symposium of Regenerative Medicine for Patients. Regen Ther. 2015;2:2-5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 82. | Hofrichter M, Nimtz L, Tigges J, Kabiri Y, Schröter F, Royer-Pokora B, Hildebrandt B, Schmuck M, Epanchintsev A, Theiss S, Adjaye J, Egly JM, Krutmann J, Fritsche E. Comparative performance analysis of human iPSC-derived and primary neural progenitor cells (NPC) grown as neurospheres in vitro. Stem Cell Res. 2017;25:72-82. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 83. | Generali M, Casanova EA, Kehl D, Wanner D, Hoerstrup SP, Cinelli P, Weber B. Autologous endothelialized small-caliber vascular grafts engineered from blood-derived induced pluripotent stem cells. Acta Biomater. 2019;97:333-343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 84. | Sundberg M, Bogetofte H, Lawson T, Jansson J, Smith G, Astradsson A, Moore M, Osborn T, Cooper O, Spealman R, Hallett P, Isacson O. Improved cell therapy protocols for Parkinson's disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells. 2013;31:1548-1562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 167] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 85. | Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1321] [Cited by in F6Publishing: 1357] [Article Influence: 150.8] [Reference Citation Analysis (1)] |
| 86. | Wang Y, Hu J, Jiao J, Liu Z, Zhou Z, Zhao C, Chang LJ, Chen YE, Ma PX, Yang B. Engineering vascular tissue with functional smooth muscle cells derived from human iPS cells and nanofibrous scaffolds. Biomaterials. 2014;35:8960-8969. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 87. | Wimmer RA, Leopoldi A, Aichinger M, Wick N, Hantusch B, Novatchkova M, Taubenschmid J, Hämmerle M, Esk C, Bagley JA, Lindenhofer D, Chen G, Boehm M, Agu CA, Yang F, Fu B, Zuber J, Knoblich JA, Kerjaschki D, Penninger JM. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 328] [Cited by in F6Publishing: 410] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 88. | Takahashi J. iPS cell-based therapy for Parkinson's disease: A Kyoto trial. Regen Ther. 2020;13:18-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 89. | Schweitzer JS, Song B, Herrington TM, Park TY, Lee N, Ko S, Jeon J, Cha Y, Kim K, Li Q, Henchcliffe C, Kaplitt M, Neff C, Rapalino O, Seo H, Lee IH, Kim J, Kim T, Petsko GA, Ritz J, Cohen BM, Kong SW, Leblanc P, Carter BS, Kim KS. Personalized iPSC-Derived Dopamine Progenitor Cells for Parkinson's Disease. N Engl J Med. 2020;382:1926-1932. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 258] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 90. | Deinsberger J, Reisinger D, Weber B. Global trends in clinical trials involving pluripotent stem cells: a systematic multi-database analysis. NPJ Regen Med. 2020;5:15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 91. | Yoshihara M, Oguchi A, Murakawa Y. Genomic Instability of iPSCs and Challenges in Their Clinical Applications. Adv Exp Med Biol. 2019;1201:23-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 92. | Mehat MS, Sundaram V, Ripamonti C, Robson AG, Smith AJ, Borooah S, Robinson M, Rosenthal AN, Innes W, Weleber RG, Lee RWJ, Crossland M, Rubin GS, Dhillon B, Steel DHW, Anglade E, Lanza RP, Ali RR, Michaelides M, Bainbridge JWB. Transplantation of Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cells in Macular Degeneration. Ophthalmology. 2018;125:1765-1775. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 147] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 93. | Liu X, Li W, Fu X, Xu Y. The Immunogenicity and Immune Tolerance of Pluripotent Stem Cell Derivatives. Front Immunol. 2017;8:645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 86] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 94. | Hermsen J, Brown ME. Humanized Mouse Models for Evaluation of PSC Immunogenicity. Curr Protoc Stem Cell Biol. 2020;54:e113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 95. | Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, Fassina A, Cordenonsi M, Piccolo S. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 697] [Cited by in F6Publishing: 725] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 96. | Nakajima-Koyama M, Lee J, Ohta S, Yamamoto T, Nishida E. Induction of Pluripotency in Astrocytes through a Neural Stem Cell-like State. J Biol Chem. 2015;290:31173-31188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 97. | Panciera T, Azzolin L, Fujimura A, Di Biagio D, Frasson C, Bresolin S, Soligo S, Basso G, Bicciato S, Rosato A, Cordenonsi M, Piccolo S. Induction of Expandable Tissue-Specific Stem/Progenitor Cells through Transient Expression of YAP/TAZ. Cell Stem Cell. 2016;19:725-737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 177] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 98. | Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783-803. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1313] [Cited by in F6Publishing: 1237] [Article Influence: 154.6] [Reference Citation Analysis (0)] |
| 99. | Marcus AJ, Woodbury D. Fetal stem cells from extra-embryonic tissues: do not discard. J Cell Mol Med. 2008;12:730-742. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 109] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 100. | O'Donoghue K, Fisk NM. Fetal stem cells. Best Pract Res Clin Obstet Gynaecol. 2004;18:853-875. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 104] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 101. | Brunstein CG, Wagner JE. Umbilical cord blood transplantation and banking. Annu Rev Med. 2006;57:403-417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 102. | Bauer G, Elsallab M, Abou-El-Enein M. Concise Review: A Comprehensive Analysis of Reported Adverse Events in Patients Receiving Unproven Stem Cell-Based Interventions. Stem Cells Transl Med. 2018;7:676-685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 103. | Wu Z, Zhang S, Zhou L, Cai J, Tan J, Gao X, Zeng Z, Li D. Thromboembolism Induced by Umbilical Cord Mesenchymal Stem Cell Infusion: A Report of Two Cases and Literature Review. Transplant Proc. 2017;49:1656-1658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 104. | Moll G, Ankrum JA, Kamhieh-Milz J, Bieback K, Ringdén O, Volk HD, Geissler S, Reinke P. Intravascular Mesenchymal Stromal/Stem Cell Therapy Product Diversification: Time for New Clinical Guidelines. Trends Mol Med. 2019;25:149-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 248] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 105. | Moll G, Ignatowicz L, Catar R, Luecht C, Sadeghi B, Hamad O, Jungebluth P, Dragun D, Schmidtchen A, Ringdén O. Different Procoagulant Activity of Therapeutic Mesenchymal Stromal Cells Derived from Bone Marrow and Placental Decidua. Stem Cells Dev. 2015;24:2269-2279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 106. | Melmed GY, Pandak WM, Casey K, Abraham B, Valentine J, Schwartz D, Awais D, Bassan I, Lichtiger S, Sands B, Hanauer S, Richards R, Oikonomou I, Parekh N, Targan S, Johnson K, Hariri R, Fischkoff S. Human Placenta-derived Cells (PDA-001) for the Treatment of Moderate-to-severe Crohn's Disease: A Phase 1b/2a Study. Inflamm Bowel Dis. 2015;21:1809-1816. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 107. | Nakamura Y, Miyaki S, Ishitobi H, Matsuyama S, Nakasa T, Kamei N, Akimoto T, Higashi Y, Ochi M. Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle regeneration. FEBS Lett. 2015;589:1257-1265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 323] [Cited by in F6Publishing: 361] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 108. | Barriga F, Ramírez P, Wietstruck A, Rojas N. Hematopoietic stem cell transplantation: clinical use and perspectives. Biol Res. 2012;45:307-316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 109. | Petersdorf EW. Optimal HLA matching in hematopoietic cell transplantation. Curr Opin Immunol. 2008;20:588-593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 109] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 110. | Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373:1550-1561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1733] [Cited by in F6Publishing: 1721] [Article Influence: 114.7] [Reference Citation Analysis (0)] |
| 111. | Dandoy CE, Kim S, Chen M, Ahn KW, Ardura MI, Brown V, Chhabra S, Diaz MA, Dvorak C, Farhadfar N, Flagg A, Ganguly S, Hale GA, Hashmi SK, Hematti P, Martino R, Nishihori T, Nusrat R, Olsson RF, Rotz SJ, Sung AD, Perales MA, Lindemans CA, Komanduri KV, Riches ML. Incidence, Risk Factors, and Outcomes of Patients Who Develop Mucosal Barrier Injury-Laboratory Confirmed Bloodstream Infections in the First 100 Days After Allogeneic Hematopoietic Stem Cell Transplant. JAMA Netw Open. 2020;3:e1918668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 112. | Cao W, Guan L, Li X, Zhang R, Li L, Zhang S, Wang C, Xie X, Jiang Z, Wan D, Chi X. Clinical Analysis of Bloodstream Infections During Agranulocytosis After Allogeneic Hematopoietic Stem Cell Transplantation. Infect Drug Resist. 2021;14:185-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 113. | Mohammadi Najafabadi M, Soleimani M, Ahmadvand M, Soufi Zomorrod M, Mousavi SA. Treatment protocols for BK virus associated hemorrhagic cystitis after hematopoietic stem cell transplantation. Am J Blood Res. 2020;10:217-230. [PubMed] [Cited in This Article: ] |
| 114. | Hass R, Kasper C, Böhm S, Jacobs R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun Signal. 2011;9:12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1018] [Cited by in F6Publishing: 1115] [Article Influence: 85.8] [Reference Citation Analysis (0)] |
| 115. | Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11055] [Cited by in F6Publishing: 11776] [Article Influence: 692.7] [Reference Citation Analysis (1)] |
| 116. | Alt E, Yan Y, Gehmert S, Song YH, Altman A, Vykoukal D, Bai X. Fibroblasts share mesenchymal phenotypes with stem cells, but lack their differentiation and colony-forming potential. Biol Cell. 2011;103:197-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 117. | Lv FJ, Tuan RS, Cheung KM, Leung VY. Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cells. 2014;32:1408-1419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 644] [Cited by in F6Publishing: 701] [Article Influence: 77.9] [Reference Citation Analysis (0)] |
| 118. | Romito A, Cobellis G. Pluripotent Stem Cells: Current Understanding and Future Directions. Stem Cells Int. 2016;2016:9451492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 119. | Golchin A, Seyedjafari E, Ardeshirylajimi A. Mesenchymal Stem Cell Therapy for COVID-19: Present or Future. Stem Cell Rev Rep. 2020;16:427-433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 207] [Article Influence: 51.8] [Reference Citation Analysis (1)] |
| 120. | Squillaro T, Peluso G, Galderisi U. Clinical Trials With Mesenchymal Stem Cells: An Update. Cell Transplant. 2016;25:829-848. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 841] [Cited by in F6Publishing: 937] [Article Influence: 104.1] [Reference Citation Analysis (0)] |
| 121. | Vizoso FJ, Eiro N, Cid S, Schneider J, Perez-Fernandez R. Mesenchymal Stem Cell Secretome: Toward Cell-Free Therapeutic Strategies in Regenerative Medicine. Int J Mol Sci. 2017;18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 548] [Cited by in F6Publishing: 713] [Article Influence: 101.9] [Reference Citation Analysis (0)] |
| 122. | Mushahary D, Spittler A, Kasper C, Weber V, Charwat V. Isolation, cultivation, and characterization of human mesenchymal stem cells. Cytometry A. 2018;93:19-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 231] [Cited by in F6Publishing: 318] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 123. | Trohatou O, Roubelakis MG. Mesenchymal Stem/Stromal Cells in Regenerative Medicine: Past, Present, and Future. Cell Reprogram. 2017;19:217-224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 124. | Galipeau J. Mesenchymal Stromal Cells for Graft-versus-Host Disease: A Trilogy. Biol Blood Marrow Transplant. 2020;26:e89-e91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 125. | Moroni L, Fornasari PM. Human mesenchymal stem cells: a bank perspective on the isolation, characterization and potential of alternative sources for the regeneration of musculoskeletal tissues. J Cell Physiol. 2013;228:680-687. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 126. | Dmitrieva RI, Minullina IR, Bilibina AA, Tarasova OV, Anisimov SV, Zaritskey AY. Bone marrow- and subcutaneous adipose tissue-derived mesenchymal stem cells: differences and similarities. Cell Cycle. 2012;11:377-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 137] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 127. | Mohamed-Ahmed S, Fristad I, Lie SA, Suliman S, Mustafa K, Vindenes H, Idris SB. Adipose-derived and bone marrow mesenchymal stem cells: a donor-matched comparison. Stem Cell Res Ther. 2018;9:168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 261] [Article Influence: 43.5] [Reference Citation Analysis (1)] |
| 128. | Iyyanki T, Hubenak J, Liu J, Chang EI, Beahm EK, Zhang Q. Harvesting technique affects adipose-derived stem cell yield. Aesthet Surg J. 2015;35:467-476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 129. | Huang SJ, Fu RH, Shyu WC, Liu SP, Jong GP, Chiu YW, Wu HS, Tsou YA, Cheng CW, Lin SZ. Adipose-derived stem cells: isolation, characterization, and differentiation potential. Cell Transplant. 2013;22:701-709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 130. | Lindroos B, Suuronen R, Miettinen S. The potential of adipose stem cells in regenerative medicine. Stem Cell Rev Rep. 2011;7:269-291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 311] [Cited by in F6Publishing: 302] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 131. | Tobita M, Orbay H, Mizuno H. Adipose-derived stem cells: current findings and future perspectives. Discov Med. 2011;11:160-170. [PubMed] [Cited in This Article: ] |
| 132. | Nombela-Arrieta C, Ritz J, Silberstein LE. The elusive nature and function of mesenchymal stem cells. Nat Rev Mol Cell Biol. 2011;12:126-131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 439] [Cited by in F6Publishing: 454] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 133. | Pittenger MF, Discher DE, Péault BM, Phinney DG, Hare JM, Caplan AI. Mesenchymal stem cell perspective: cell biology to clinical progress. NPJ Regen Med. 2019;4:22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 773] [Cited by in F6Publishing: 937] [Article Influence: 187.4] [Reference Citation Analysis (0)] |
| 134. | Ren G, Rezaee M, Razavi M, Taysir A, Wang J, Thakor AS. Adipose tissue-derived mesenchymal stem cells rescue the function of islets transplanted in sub-therapeutic numbers via their angiogenic properties. Cell Tissue Res. 2019;376:353-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 135. | Le Blanc K, Rasmusson I, Sundberg B, Götherström C, Hassan M, Uzunel M, Ringdén O. Treatment of severe acute graft-versus-host disease with third party haploidentical mesenchymal stem cells. Lancet. 2004;363:1439-1441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2044] [Cited by in F6Publishing: 1970] [Article Influence: 98.5] [Reference Citation Analysis (0)] |
| 136. | Moll G, Drzeniek N, Kamhieh-Milz J, Geissler S, Volk HD, Reinke P. MSC Therapies for COVID-19: Importance of Patient Coagulopathy, Thromboprophylaxis, Cell Product Quality and Mode of Delivery for Treatment Safety and Efficacy. Front Immunol. 2020;11:1091. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 137. | Wu X, Jiang J, Gu Z, Zhang J, Chen Y, Liu X. Mesenchymal stromal cell therapies: immunomodulatory properties and clinical progress. Stem Cell Res Ther. 2020;11:345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 129] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 138. | Kabat M, Bobkov I, Kumar S, Grumet M. Trends in mesenchymal stem cell clinical trials 2004-2018: Is efficacy optimal in a narrow dose range? Stem Cells Transl Med. 2020;9:17-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 248] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 139. | Kurtzberg J, Abdel-Azim H, Carpenter P, Chaudhury S, Horn B, Mahadeo K, Nemecek E, Neudorf S, Prasad V, Prockop S, Quigg T, Satwani P, Cheng A, Burke E, Hayes J, Skerrett D; MSB-GVHD001/002 Study Group. A Phase 3, Single-Arm, Prospective Study of Remestemcel-L, Ex Vivo Culture-Expanded Adult Human Mesenchymal Stromal Cells for the Treatment of Pediatric Patients Who Failed to Respond to Steroid Treatment for Acute Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2020;26:845-854. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 84] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 140. | Kurtzberg J, Prockop S, Chaudhury S, Horn B, Nemecek E, Prasad V, Satwani P, Teira P, Hayes J, Burke E; MSB-275 Study Group. Study 275: Updated Expanded Access Program for Remestemcel-L in Steroid-Refractory Acute Graft-versus-Host Disease in Children. Biol Blood Marrow Transplant. 2020;26:855-864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 141. | Kebriaei P, Hayes J, Daly A, Uberti J, Marks DI, Soiffer R, Waller EK, Burke E, Skerrett D, Shpall E, Martin PJ. A Phase 3 Randomized Study of Remestemcel-L vs Placebo Added to Second-Line Therapy in Patients with Steroid-Refractory Acute Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2020;26:835-844. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 142. | Panés J, García-Olmo D, Van Assche G, Colombel JF, Reinisch W, Baumgart DC, Dignass A, Nachury M, Ferrante M, Kazemi-Shirazi L, Grimaud JC, de la Portilla F, Goldin E, Richard MP, Leselbaum A, Danese S; ADMIRE CD Study Group Collaborators. Expanded allogeneic adipose-derived mesenchymal stem cells (Cx601) for complex perianal fistulas in Crohn's disease: a phase 3 randomised, double-blind controlled trial. Lancet. 2016;388:1281-1290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 628] [Cited by in F6Publishing: 624] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 143. | Panés J, García-Olmo D, Van Assche G, Colombel JF, Reinisch W, Baumgart DC, Dignass A, Nachury M, Ferrante M, Kazemi-Shirazi L, Grimaud JC, de la Portilla F, Goldin E, Richard MP, Diez MC, Tagarro I, Leselbaum A, Danese S; ADMIRE CD Study Group Collaborators. Long-term Efficacy and Safety of Stem Cell Therapy (Cx601) for Complex Perianal Fistulas in Patients With Crohn's Disease. Gastroenterology 2018; 154: 1334-1342. e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 275] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 144. | Karantalis V, Hare JM. Use of mesenchymal stem cells for therapy of cardiac disease. Circ Res. 2015;116:1413-1430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 303] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 145. | Bartunek J, Davison B, Sherman W, Povsic T, Henry TD, Gersh B, Metra M, Filippatos G, Hajjar R, Behfar A, Homsy C, Cotter G, Wijns W, Tendera M, Terzic A. Congestive Heart Failure Cardiopoietic Regenerative Therapy (CHART-1) trial design. Eur J Heart Fail. 2016;18:160-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 146. | Teerlink JR, Metra M, Filippatos GS, Davison BA, Bartunek J, Terzic A, Gersh BJ, Povsic TJ, Henry TD, Alexandre B, Homsy C, Edwards C, Seron A, Wijns W, Cotter G; CHART Investigators. Benefit of cardiopoietic mesenchymal stem cell therapy on left ventricular remodelling: results from the Congestive Heart Failure Cardiopoietic Regenerative Therapy (CHART-1) study. Eur J Heart Fail. 2017;19:1520-1529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 147. | Wang D, Zhang H, Liang J, Li X, Feng X, Wang H, Hua B, Liu B, Lu L, Gilkeson GS, Silver RM, Chen W, Shi S, Sun L. Allogeneic mesenchymal stem cell transplantation in severe and refractory systemic lupus erythematosus: 4 years of experience. Cell Transplant. 2013;22:2267-2277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 171] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 148. | Wang D, Zhang H, Liang J, Wang H, Hua B, Feng X, Gilkeson GS, Farge D, Shi S, Sun L. A Long-Term Follow-Up Study of Allogeneic Mesenchymal Stem/Stromal Cell Transplantation in Patients with Drug-Resistant Systemic Lupus Erythematosus. Stem Cell Reports. 2018;10:933-941. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 149. | Cai J, Wu Z, Xu X, Liao L, Chen J, Huang L, Wu W, Luo F, Wu C, Pugliese A, Pileggi A, Ricordi C, Tan J. Umbilical Cord Mesenchymal Stromal Cell With Autologous Bone Marrow Cell Transplantation in Established Type 1 Diabetes: A Pilot Randomized Controlled Open-Label Clinical Study to Assess Safety and Impact on Insulin Secretion. Diabetes Care. 2016;39:149-157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 113] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 150. | Fričová D, Korchak JA, Zubair AC. Challenges and translational considerations of mesenchymal stem/stromal cell therapy for Parkinson's disease. NPJ Regen Med. 2020;5:20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 151. | Liu XY, Yang LP, Zhao L. Stem cell therapy for Alzheimer's disease. World J Stem Cells. 2020;12:787-802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 57] [Article Influence: 14.3] [Reference Citation Analysis (10)] |
| 152. | Cell Trials Data. The Role of MSC to Treat Coronavirus Pneumonia and ARDS. Part 1: Is the Emperor Wearing Clothes? [cited 20 Jan 2021]. In: Cell Trials Data [Internet]. Available online at: https://celltrials.org/news/role-msc-treat-coronavirus-pneumoniaand-ards-part-1-is-emperor-wearing-clothes. [Cited in This Article: ] |
| 153. | Kumar R, Sharma A, Siddiqui MH, Tiwari RK. Prediction of Metabolism of Drugs using Artificial Intelligence: How far have we reached? Curr Drug Metab. 2016;17:129-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 154. | Kumar R, Sharma A, Siddiqui MH, Tiwari RK. Prediction of Human Intestinal Absorption of Compounds Using Artificial Intelligence Techniques. Curr Drug Discov Technol. 2017;14:244-254. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 155. | Kumar R, Sharma A, Siddiqui MH, Tiwari RK. Promises of Machine Learning Approaches in Prediction of Absorption of Compounds. Mini Rev Med Chem. 2018;18:196-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 156. | Haenlein M, Kaplan A. A brief history of artificial intelligence: On the past, present, and future of artificial intelligence. Calif Manage Rev. 2019;61:5-14. [DOI] [Cited in This Article: ] |
| 157. | Wang L, Zhang HC, Wang Q. On the concepts of artificial intelligence and innovative design in product design. IOP Conf Ser Mater Sci Eng. 2019;573:12095. [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 158. | Kumar R, Sharma A, Varadwaj P, Ahmad A, Ashraf GM. Classification of oral bioavailability of drugs by machine learning approaches: a comparative study. J Comp Int Sci. 2011;2:1-18. [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 159. | He J, Baxter SL, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med. 2019;25:30-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 597] [Cited by in F6Publishing: 673] [Article Influence: 134.6] [Reference Citation Analysis (0)] |
| 160. | Kumar R, Sharma A, Siddiqui MH, Tiwari RK. Prediction of Drug-Plasma Protein Binding Using Artificial Intelligence Based Algorithms. Comb Chem High Throughput Screen. 2018;21:57-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 161. | Sharma A, Kumar R, Ranjta S, Varadwaj PK. SMILES to Smell: Decoding the Structure-Odor Relationship of Chemical Compounds Using the Deep Neural Network Approach. J Chem Inf Model. 2021;61:676-688. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 32] [Article Influence: 10.7] [Reference Citation Analysis (1)] |
| 162. | Jiang F, Jiang Y, Zhi H, Dong Y, Li H, Ma S, Wang Y, Dong Q, Shen H. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2017;2:230-243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1189] [Cited by in F6Publishing: 1014] [Article Influence: 144.9] [Reference Citation Analysis (0)] |
| 163. | Buch VH, Ahmed I, Maruthappu M. Artificial intelligence in medicine: current trends and future possibilities. Br J Gen Pract. 2018;68:143-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 108] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 164. | Salehnasab C, Hajifathali A, Asadi F, Roshandel E, Kazemi A, Roshanpoor A. Machine Learning Classification Algorithms to Predict aGvHD following Allo-HSCT: A Systematic Review. Methods Inf Med. 2019;58:205-212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 165. | Radakovich N, Nagy M, Nazha A. Artificial Intelligence in Hematology: Current Challenges and Opportunities. Curr Hematol Malig Rep. 2020;15:203-210. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 166. | Saba L, Biswas M, Kuppili V, Cuadrado Godia E, Suri HS, Edla DR, Omerzu T, Laird JR, Khanna NN, Mavrogeni S, Protogerou A, Sfikakis PP, Viswanathan V, Kitas GD, Nicolaides A, Gupta A, Suri JS. The present and future of deep learning in radiology. Eur J Radiol. 2019;114:14-24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 154] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 167. | Kusumoto D, Yuasa S. The application of convolutional neural network to stem cell biology. Inflamm Regen. 2019;39:14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 168. | Joutsijoki H, Haponen M, Rasku J, Aalto-Setälä K, Juhola M. Machine Learning Approach to Automated Quality Identification of Human Induced Pluripotent Stem Cell Colony Images. Comput Math Methods Med. 2016;2016:3091039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 169. | Kavitha MS, Kurita T, Ahn BC. Critical texture pattern feature assessment for characterizing colonies of induced pluripotent stem cells through machine learning techniques. Comput Biol Med. 2018;94:55-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 170. | Kavitha MS, Kurita T, Park SY, Chien SI, Bae JS, Ahn BC. Deep vector-based convolutional neural network approach for automatic recognition of colonies of induced pluripotent stem cells. PLoS One. 2017;12:e0189974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 171. | Kusumoto D, Lachmann M, Kunihiro T, Yuasa S, Kishino Y, Kimura M, Katsuki T, Itoh S, Seki T, Fukuda K. Automated Deep Learning-Based System to Identify Endothelial Cells Derived from Induced Pluripotent Stem Cells. Stem Cell Reports. 2018;10:1687-1695. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 172. | Waisman A, La Greca A, Möbbs AM, Scarafía MA, Santín Velazque NL, Neiman G, Moro LN, Luzzani C, Sevlever GE, Guberman AS, Miriuka SG. Deep Learning Neural Networks Highly Predict Very Early Onset of Pluripotent Stem Cell Differentiation. Stem Cell Reports. 2019;12:845-859. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 173. | Juhola M, Penttinen K, Joutsijoki H, Aalto-Setälä K. Analysis of Drug Effects on iPSC Cardiomyocytes with Machine Learning. Ann Biomed Eng. 2021;49:129-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 174. | Juhola M, Penttinen K, Joutsijoki H, Varpa K, Saarikoski J, Rasku J, Siirtola H, Iltanen K, Laurikkala J, Hyyrö H, Hyttinen J, Aalto-Setälä K. Signal analysis and classification methods for the calcium transient data of stem cell-derived cardiomyocytes. Comput Biol Med. 2015;61:1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 175. | Hwang H, Liu R, Maxwell JT, Yang J, Xu C. Machine learning identifies abnormal Ca2+ transients in human induced pluripotent stem cell-derived cardiomyocytes. Sci Rep. 2020;10:16977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 176. | Nishino K, Takasawa K, Okamura K, Arai Y, Sekiya A, Akutsu H, Umezawa A. Identification of an epigenetic signature in human induced pluripotent stem cells using a linear machine learning model. Hum Cell. 2021;34:99-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 177. | Liu YYF, Lu Y, Oh S, Conduit GJ. Machine learning to predict mesenchymal stem cell efficacy for cartilage repair. PLoS Comput Biol. 2020;16:e1008275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 178. | Lee LX, Li SC. Hunting down the dominating subclone of cancer stem cells as a potential new therapeutic target in multiple myeloma: An artificial intelligence perspective. World J Stem Cells. 2020;12:706-720. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 179. | Ng AP, Alexander WS. Haematopoietic stem cells: past, present and future. Cell Death Discov. 2017;3:17002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 180. | Sniecinski I, Seghatchian J. Artificial intelligence: A joint narrative on potential use in pediatric stem and immune cell therapies and regenerative medicine. Transfus Apher Sci. 2018;57:422-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 181. | Polak DJ. Regenerative medicine. Opportunities and challenges: a brief overview. J R Soc Interface. 2010;7 Suppl 6:S777-S781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 182. | Durant F, Lobo D, Hammelman J, Levin M. Physiological controls of large-scale patterning in planarian regeneration: a molecular and computational perspective on growth and form. Regeneration (Oxf). 2016;3:78-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 183. | Roost MS, van Iperen L, Ariyurek Y, Buermans HP, Arindrarto W, Devalla HD, Passier R, Mummery CL, Carlotti F, de Koning EJ, van Zwet EW, Goeman JJ, Chuva de Sousa Lopes SM. KeyGenes, a Tool to Probe Tissue Differentiation Using a Human Fetal Transcriptional Atlas. Stem Cell Reports. 2015;4:1112-1124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 184. | Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158:889-902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 194] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 185. | Shamblott MJ, Axelman J, Wang S, Bugg EM, Littlefield JW, Donovan PJ, Blumenthal PD, Huggins GR, Gearhart JD. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc Natl Acad Sci USA. 1998;95:13726-13731. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1002] [Cited by in F6Publishing: 842] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 186. | Lorenz E, Uphoff D, Reid Tr, Shelton E. Modification of irradiation injury in mice and guinea pigs by bone marrow injections. J Natl Cancer Inst. 1951;12:197-201. [PubMed] [Cited in This Article: ] |
| 187. | Thomas ED, Storb R, Fefer A, Slichter SJ, Bryant JI, Buckner CD, Neiman PE, Clift RA, Funk DD, Lerner KE. Aplastic anaemia treated by marrow transplantation. Lancet. 1972;1:284-289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 173] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 188. | Baron JH. Evolution of clinical research: A history before and beyond James Lind. Perspect Clin Res. 2012;3:149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |