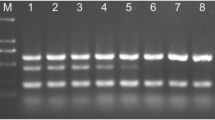
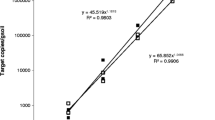
Abstract
In this study, we report on the development of two quantitative PCR assays for the detection and quantification of the soilborne plant pathogen Phytophthora cactorum causing root and crown rot of strawberry. The assays rely on the use of SYBR Green I chemistry and use the internal transcribed spacer region 2 (ITS2) and the Ras-related protein gene Ypt1 as detection markers. An extensive list of Phytophthora isolates was included in the study to evaluate assay specificity. High specificity was obtained when Ypt1 was used as detection marker, with cross-reactions only occurring with the closest relatives of P. cactorum, including P. hedraiandra, P. idaei and P. pseudotsugae. In contrast, the ITS-based assay was less specific, resulting in a larger number of cross-reactions (eight species when tested at 10 pg DNA). In sensitivity tests, P. cactorum DNA was detected down to 10 fg using the ITS-based assay, while the limit of detection (LOD) for the Ypt1-based assay was 1 pg DNA, irrespective of the presence of background DNA from strawberry or soil. When strawberry, growth substrate and soil samples were spiked with a 10-fold dilution series of P. cactorum zoospores, the LOD for the ITS-based assay was 10 zoospores g−1 plant material, growth substrate or soil. The limit of quantification (LOQ) of the assay was 109 zoospores g−1 plant material, 15.8 zoospores g−1 growth substrate, and 106 zoospores g−1 soil. For the Ypt1-based assay, the LOD and LOQ values were 1000 and 5910 zoospores g−1 plant material, 1000 and 1818 zoospores g−1 growth substrate, and 1000 and 3823 zoospores g−1 soil, respectively. In terms of detection, analysis of field samples suggested that the Ypt1-based assay almost performed equally well compared to the ITS-based assay, especially for plant and growth substrate samples. Further, our results showed that both qPCR assays outperformed classical plating, illustrating their power to be used for diagnostic purposes. Interestingly, P. cactorum was also detected in soil and growth substrate samples from areas with no symptoms, suggesting that the assays can also aid in early pathogen detection before the disease manifests.



Similar content being viewed by others
Data availability
Not applicable.
Code availability
Not applicable.
References
Akino, S., Takemoto, D., & Hosaka, K. (2014). Phytophthora infestans: A review of past and current studies on potato late blight. Journal of General Plant Pathology, 80, 24–37.
Barboza, E. A., Fonseca, M. E. N., Boiteux, L. S., & Reis, A. (2017). First worldwide report of a strawberry fruit rot disease caused by Phytophthora capsici isolates. Plant Disease, 101, 259–260.
Blair, J. E., Coffey, M. D., Park, S. Y., Geiser, D. M., & Kang, S. (2008). A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology, 45, 266–277.
Bonants, P. J. M., Hagenaar-de Weerdt, M., Man In ‘t Veld, W. A., & Baayen, R. P. (2000). Molecular characterization of natural hybrids of Phytophthora nicotianae and P. cactorum. Phytopathology, 90, 867–874.
Capote, N., Pastrana, A. M., Aguado, A., & Sanchez-Torres, P. (2012). Molecular tools for detection of plant pathogenic fungi and fungicide resistance. In Dr. C. J. Cumagun (Ed.), Plant pathology (pp. 151–202). London: IntechOpen.
Catal, M., Erler, F., Fulbright, D. W., & Adams, G. C. (2013). Real-time quantitative PCR assays for evaluation of soybean varieties for resistance to the stem and root rot pathogen Phytophthora sojae. European Journal of Plant Pathology, 137, 859–869.
Chen, Y., & Roxby, R. (1996). Characterization of a Phytophthora infestans gene involved in vesicle transport. Gene, 181, 89–94.
Chimento, A., Cacciola, S. O., & Garbelotto, M. (2011). Detection of mRNA by reverse-transcription PCR as an indicator of viability in Phytophthora ramorum. Forest Pathology, 42, 14–21.
Cooke, D. E. L., Schena, L., & Cacciola, S. O. (2007). Tools to detect, identify and monitor Phytophthora species in natural ecosystems. Journal of Plant Pathology, 89, 13–28.
Davidson, J. M., Werres, S., Garbelotto, M., Hansen, E. M., & Rizzo, D. M. (2003). Sudden oak death and associated diseases caused by Phytophthora ramorum. Plant Health Progress, 4, 12. https://doi.org/10.1094/PHP-2003-0707-01-DG.
Dolezel, J., Bartos, J., Voglmayr, H., & Greilhuber, J. (2003). Nuclear DNA content and genome size of trout and human. Cytometry. Part A: The Journal of the International Society for Analytical Cytology, 51, 127–128.
Ellis, M. A., & Grove, G. G. (1983). Leather rot in Ohio strawberries. Plant Disease, 67, 549.
Engelberg, J., Duong, T. A., & van den Berg, N. (2013). Development of a nested quantitative real-time PCR for detecting Phytophthora cinnamomi in Persea americana rootstocks. Plant Disease, 97, 1012–1017.
Englander, L., & Roth, L. F. (1979). Interaction of light and sterol on sporangium and chlamydospore production by Phytophthora lateralis. Phytopathology, 70, 650–654.
Erwin, D. C., & Ribeiro, O. K. (1996). Phytophthora Diseases Worldwide. St. Paul, MN: APS Press.
Eshraghi, L., Aryamanesh, N., Anderson, J. P., Shearer, B., McComb, J. A., Hardy, G. E. S. J., & O’Brien, P. A. (2011). A quantitative PCR assay for accurate in planta quantification of the necrotrophic pathogen Phytophthora cinnamomi. European Journal of Plant Pathology, 131, 419–430.
Faedda, R., Cacciola, S. O., Pane, A., Szigethy, A., Bakonyi, J., Man In’t Veld, W. A., et al. (2013). Phytophthora × pelgrandis causes root and collar rot of Lavandula stoechas in Italy. Plant Disease, 97, 1091–1096.
Ferguson, A. J., & Jeffers, S. N. (1999). Detecting multiple species of Phytophthora in container mixes from ornamental crop nurseries. Plant Disease, 83, 1129–1136.
Fittipaldi, M., Nocker, A., & Codony, F. (2012). Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. Journal of Microbiological Methods, 91, 276–289.
Forootan, A., Sjöback, R., Björkman, J., Sjögreen, B., Linz, L., & Kubista, M. (2017). Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomolecular Detection and Quantification, 12, 1–6.
French-Monar, R. D., Jones, J. B., Ozores-Hampton, M., & Roberts, P. D. (2007). Survival of inoculum of Phytophthora capsici in soil through time under different soil treatments. Plant Disease, 91, 593–598.
Garrido, C., Carbú, M., Fernández-Acero, F. J., González-Rodríguez, V. E., & Cantoral, J. M. (2011). New insights in the study of strawberry fungal pathogens. Genes, Genomes and Genomics, 5, 24–39.
Garrido, C., González-Rodríguez, V. E., Carbú, M., Husaini, A. M., & Cantoral, J. M. (2016). Fungal diseases of strawberry and their diagnosis. In A. M. Husaini & D. Neri (Eds.), Strawberry: Growth, development and diseases (pp. 157–184). Boston: CAB International.
Harris, D. C. (1986). Methods for preparing, estimating and diluting suspensions of Phytophthora cactorum zoospores. Transactions of the British Mycological Society, 86, 482–486.
Heise, J., Nega, M., Alawi, M., & Wagner, D. (2016). Propidium monoazide treatment to distinguish between live and dead methanogens in pure cultures and environmental samples. Journal of Microbiological Methods, 121, 11–23.
Judelson, H. S., & Blanco, F. A. (2005). The spores of Phytophthora: Weapons of the plant destroyer. Nature Reviews Microbiology, 3, 47–58.
Khan, M., Li, B., Jiang, Y., Weng, Q., & Chen, Q. (2017). Evaluation of different PCR-based assays and LAMP method for rapid detection of Phytophthora infestans by targeting the Ypt1 gene. Frontiers in Microbiology, 8, 1920.
König, S., Schwenkbier, L., Pollok, S., Riedel, M., Wagner, S., Popp, J., Weber, K., & Werres, S. (2015). Potential of Ypt1 and ITS gene regions for the detection of Phytophthora species in a lab-on-a-chip DNA hybridization array. Plant Pathology, 64, 1176–1189.
Kroon, L. P. N. M., Bakker, F. T., van den Bosch, G. B. M., Bonants, P. J. M., & Flier, W. G. (2004). Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genetics and Biology, 41, 766–782.
Kunadiya, M. B., Dunstan, W. D., White, D., Hardy, G. E. S. J., Grigg, A. H., & Burgess, T. I. (2019). A qPCR assay for the detection of Phytophthora cinnamomi including an mRNA protocol designed to establish propagule viability in environmental samples. Plant Disease, 103, 2443–2450.
Lees, A. K., Sullivan, L., Lynott, J. S., & Cullen, D. W. (2012). Development of a quantitative real-time PCR assay for Phytophthora infestans and its applicability to leaf, tuber and soil samples. Plant Pathology, 61, 867–876.
Li, M., Inada, M., Watanabe, H., Suga, H., & Kageyama, K. (2013). Simultaneous detection and quantification of Phytophthora nicotianae and P. cactorum, and distribution analyses in strawberry greenhouses by duplex real-time PCR. Microbes and Environments, 28, 195–203.
Lievens, B., Brouwer, M., Vanachter, A. C. R. C., Lévesque, C. A., Cammue, B. P. A., & Thomma, B. P. H. J. (2003). Design and development of a DNA array for rapid detection and identification of multiple tomato vascular wilt pathogens. FEMS Microbiology Letters, 223, 113–122.
Lievens, B., Grauwet, T. J. M. A., Cammue, B. P. A., & Thomma, B. P. H. J. (2005). Recent developments in diagnostics of plant pathogens: A review. In S. G. Pandalai (Ed.), Recent research developments in microbiology (pp. 57–79). Kerala, India.
Man In’t Veld, W. A., Rosendahl, K. C. H. M., & Hong, C. (2012). Phytophthora x serendipita sp. nov. and P. x pelgrandis, two destructive pathogens generated by natural hybridization. Mycologia, 104, 1390–1396.
Martin, F. N., & Tooley, P. W. (2003). Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia, 95, 269–284.
Martin, F., Abad, Z. G., Balci, Y., & Ivors, K. (2012). Identification and detection of Phytophthora: Reviewing our progress, identifying our needs. Plant Disease, 96, 1080–1103.
Mauvisseau, Q., Burian, A., Gibson, C., Brys, R., Ramsey, A., & Sweet, M. (2019). Influence of accuracy, repeatability and detection probability in the reliability of species-specific eDNA based approaches. Scientific Reports, 9, 580.
McIntosh, D. L. (1977). Phytophthora cactorum propagule density levels in orchard soil. The Plant Disease Reporter, 61, 528–532.
Nowakowska, J. A., Malewski, T., Tereba, A., & Oszako, T. (2017). Rapid diagnosis of pathogenic Phytophthora species in soil by real-time PCR. Forest Pathology. https://doi.org/10.1111/efp.12303.
O’Brien, P. A., Williams, N., & Hardy, G. E. S. (2009). Detecting Phytophthora. Critical Reviews in Microbiology, 35, 169–181.
Pánek, M., Fér, T., Mráček, J., & Tomšovský, M. (2016). Evolutionary relationships within the Phytophthora cactorum species complex in Europe. Fungal Biology, 120, 836–851.
Pietramellara, G., Ascher, J., Borgogni, F., Ceccherini, M. T., Guerri, G., & Nannipieri, P. (2009). Extracellular DNA in soil and sediment: Fate and ecological relevance. Biology and Fertility of Soils, 45, 219–235.
Porras, M., Barrau, C., Arroyo, F. T., Santos, B., Blanco, C., & Romero, F. (2007). Reduction of Phytophthora cactorum in strawberry fields by Trichoderma spp. and soil solarization. Plant Disease, 91, 142–146.
R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL: http://www.r-project.org/index.html.
Rozen, S., & Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. In S. Misener & S. A. Krawetz (Eds.), Bioinformatics methods and protocols (pp. 365–386). Totawa, NJ: Humana Press.
Schena, L., & Cooke, D. E. L. (2006). Assessing the potential of regions of the nuclear and mitochondrial genome to develop a “molecular tool box” for the detection and characterization of Phytophthora species. Journal of Microbiological Methods, 67, 70–85.
Schena, L., Nigro, F., Ippolito, A., & Gallitelli, D. (2004). Real-time quantitative PCR: A new technology to detect and study phytopathogenic and antagonistic fungi. European Journal of Plant Pathology, 110, 893–908.
Schena, L., Duncan, J. M., & Cooke, D. E. L. (2008). Development and application of a PCR-based ‘molecular tool box’ for the identification of Phytophthora species damaging forests and natural ecosystems. Plant Pathology, 57, 64–75.
Schena, L., Li Destri Nicosia, M. G., Sanzani, S. M., Faedda, R., Ippolito, A., & Cacciola, S. O. (2013). Development of quantitative PCR detection methods for phytopathogenic fungi and oomycetes. Journal of Plant Pathology, 95, 7–24.
Scheu, P. M., Berghof, K., & Stahl, U. (1998). Detection of pathogenic and spoilage micro-organisms in food with the polymerase chain reaction. Food Microbiology, 15, 13–31.
Stensvand, A., Herrero, M. L., & Talgø, V. (1999). Crown rot caused by Phytophthora cactorum in Norwegian strawberry production. EPPO Bulletin, 29, 255–158.
Storts, D. R. (2014). Alternative probe-based detection systems in quantitative PCR. The Journal of Molecular Diagnostics, 16, 614.
Tajadini, M., Panjehpour, M., & Javanmard, S. H. (2014). Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Advanced Biomedical Research, 3, 85.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Wagner, A. O., Malin, C., Knapp, B. A., & Illmer, P. (2008). Removal of free extracellular DNA from environmental samples by ethidium monoazide and propidium monoazide. Applied and Environmental Microbiology, 74, 2537–2539.
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M. A., Gelfand, D. H., Sninsky, J. J. & White, T. J. (Eds.), PCR Protocols. A guide to methods and applications. (pp. 315–222). New York: Academic Press, Inc.
Yang, X., Tyler, B. M., & Hong, C. (2017). An expanded phylogeny for the genus Phytophthora. IMA Fungus, 8, 355–384.
Yang, M., Duan, S., Mei, X., Huang, H., Chen, W., Liu, Y., Guo, C., Yang, T., Wei, W., Liu, X., He, X., Dong, Y., & Zhu, S. (2018). The Phytophthora cactorum genome provides insights into the adaptation to host defense compounds and fungicides. Scientific Reports, 8, 6534.
Acknowledgments
We would like to thank VLAIO (Flanders Innovation and Entrepreneurship) for financial support (HBC.2017.0833). Additionally, we are grateful to our colleagues W. Man in ‘t Veld, A. Numansen, L. Kroon, W. Flier, F. Govers, D. Cooke, K. Hughes, S. Werres, J. Bakyoni, S. Klemsdal, E. Hansen and M. Coffey for giving us access their Phytophthora collections. Further, we would like to thank all growers providing samples for this research.
Funding
This study was funded by VLAIO (Flanders Innovation and Entrepreneurship), project HBC.2017.0833.
Author information
Authors and Affiliations
Contributions
EV, HR and BL conceived the ideas and designed methodology. EV, AC, DB and JF collected the data. PM, WVH, HR and BL contributed to infrastructure, equipment, and reagents. PB provided DNA samples. EV, HR and BL analyzed the data. EV and BL led the writing of the manuscript. All authors contributed critically to the drafts of this manuscript and gave final approval for publication.
Corresponding author
Ethics declarations
Ethics approval
All authors gave approval for submission of this manuscript. The manuscript has been prepared following principles of ethical and professional conduct. The research did not involve human participants or animals. Therefore, neither statements concerning informed consent nor welfare of animals is applicable.
Additional declarations for articles in life science journals that report the results of studies involving humans and/or animals
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Consent for publication was obtained from all co-authors.
Conflicts of interest/competing interests
The authors declare that no conflicts of interest exist.
Additional information
Rediers, H. and Lievens, B. are shared last co-author
Supplementary Information
ESM 1
(DOCX 147 kb)
Rights and permissions
About this article
Cite this article
Verdecchia, E., Ceustermans, A., Baets, D. et al. Quantitative PCR for detection and quantification of Phytophthora cactorum in the cultivation of strawberry. Eur J Plant Pathol 160, 867–882 (2021). https://doi.org/10.1007/s10658-021-02290-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-021-02290-z