Abstract
Wheat (Triticum aestivum L.) yields are commonly affected by foliar infection by fungal pathogens. Of these, three wheat leaf blotch fungal diseases, septoria nodorum blotch (SNB), tan spot (TS) and septoria tritici blotch (STB), caused by Parastagonospora nodorum (Pn), Pyrenophora tritici-repentis (Ptr) and Zymoseptoria tritici (Zt), respectively, induce major yield losses. Infection results in necrotic areas on the leaf, and it is often difficult to determine the underlying causative pathogen from visible symptoms alone, especially in mixed infections. Here, a regional survey of 330 wheat samples collected across three seasons (years 2015–2017) from four north-west European countries was undertaken. Using quantitative polymerase chain reaction (qPCR) assays specific for each pathogen, as well as disease assessment of leaf materials, distinct regional differences were identified. Two-thirds (65%) of all samples harbored at least two of the three pathogens. Norway had high SNB abundance, but also showed mixed infections of SNB, TS and STB. In Germany, TS was prevalent, with STB also common. Danish samples commonly possessed all three pathogens, with STB prevalent, followed by TS and SNB. The UK had a major prevalence of STB with minimal occurrence of TS and SNB. Across all samples, qPCR identified Zt, Pn and Ptr in 90%, 54% and 57% of samples, respectively. For each pathogen, average disease levels via visual assessment showed modest positive correlation with fungal DNA concentrations (R2 = 0.13–0.32). Overall, our study highlights that the occurrence of mixed infection is common and widespread, with important implications for wheat disease management and breeding strategies.
Similar content being viewed by others
Introduction
Wheat (Triticum aestivum L.) is a crop of global importance, with the European Union (EU) representing the world’s highest producer. It is a major food and feed crop in Europe, is a global commodity, and plays a critical role in supporting worldwide food security. Three leaf blotch diseases of wheat caused by fungal pathogens induce major yield losses in Europe and worldwide (Singh et al., 2016): septoria nodorum blotch (SNB), caused by Parastagonospora nodorum (Pn); tan spot (TS), caused by Pyrenophora tritici-repentis (Ptr); and septoria tritici blotch (STB), caused by Zymoseptoria tritici (Zt). All three diseases result in chlorotic spots on leaves which later develop into necrotic lesions, appearing first on the lower leaves and progressing upwards during the growing season via physical dispersal mechanisms such as rain splash (Abdullah et al., 2018; Serfling et al., 2017). The similarity in symptoms means it is often difficult to distinguish these diseases visually, especially when mixed infections occur. Indeed, often no attempt is made during field assessment to discriminate within a disease complex. Recently, the increased importance of these three diseases has been linked to various factors, such as intensified cropping of wheat, shorter crop rotations, monoculture, and increased area with conservation farming (minimal tillage) (Singh et al., 2016).
All three diseases are undergoing changes in their European distributions. STB is heavily researched (e.g. Fones & Gurr, 2015), while SNB and TS have been less intensively studied, and their occurrence and degree of crop losses are less understood. Although Ptr was isolated and characterized in the early 1900s, TS in wheat did not become a serious problem until the 1970s (in the USA, De Wolf et al., 1988) and is now found in many countries worldwide (Moreno et al., 2012). In Europe, Ptr is known to be a major pathogen in regions of Germany and Denmark, mainly linked to reduced tillage and intensive wheat cropping (Jørgensen & Olsen, 2007). Until the 1960s, SNB was the major wheat disease in north-western Europe but has since been largely replaced by STB (Bayles, 1991; Shaw, 1999). SNB decline has been variously attributed to increased growing of STB-susceptible varieties, differential response to fungicides, increased fertilizer use, climate change, and a decline in atmospheric SO2 (Bearchell et al., 2005; Shaw et al., 2008; West et al., 2012).
To date, few investigations have documented the extent to which the three diseases co-exist (e.g. Blixt et al., 2010; Sapkota et al., 2015). This is assumed to largely be due to the difficulty in distinguishing them visually in mixed infection, requiring alternative methods such as incubating leaf sections to encourage sporulation (e.g. Krupinsky & Berdahl, 1984). For example, Loughman et al. (1994) found that disease symptoms in the field under mixed infection by all three pathogens prevented accurate determination of variety resistance, and that investigation of the proportion of leaf area sporulating with each pathogen was required to determine varietal response.
Virulence in Ptr and Pn is controlled, at least in part, by the interaction of necrotrophic effectors and host recognition loci. Within this system, the amount of disease represents a function of the number and activity of effectors expressed by the pathogen for which matching effector sensitivity alleles are present in the wheat plant. This is known as effector-triggered susceptibility and is genetically characterized via an `inverse gene for gene system’ (Friesen et al., 2007). Effector sensitivities are predominantly controlled by single major genetic loci, and so can be easily tracked and manipulated by breeders and researchers. Four effectors were cloned and characterized for their role in TS and SNB: ToxA (present in both Ptr and Pn), ToxB (Ptr), Tox1 (Pn) and Tox3 (Pn) (Faris et al., 2013; Friesen et al., 2007; reviewed by Downie et al., 2020). These four effectors explain a significant fraction of SNB and/or TS disease levels in agricultural environments as different as Australia (Vleeshouwers & Oliver, 2014) and Norway (Lin et al., 2020; Ruud et al., 2017), and there are clearly more to be found. For STB, many resistance QTL have been reported (e.g. Brown et al., 2015; Kollers et al., 2013; Stadlmeier et al., 2019). The recent cloning of the Z. tritici effector AvrStb6 (Zhong et al., 2017), as well as the corresponding wheat sensitivity locus Stb6 (Saintenac et al., 2018), represent the first components of gene-for-gene interaction to be functionally characterised in the Z. tritici-wheat pathosystem.
Despite breeding efforts to date, high-yielding European cultivars generally possess only moderate resistance to these three leaf blotch diseases (Singh et al., 2016; HGCA, 2021). Therefore, control of the three necrotrophic fungi by fungicides has been common practice. However, problems with fungicide resistance and stricter regulations in Europe make future control of these pathogens by fungicides more uncertain. Already, STB control is compromised by resistance to several major fungicide classes (Cools & Fraaije, 2013; Fraaije et al., 2005; Rehfus et al., 2017; Torriani et al., 2009), and TS and SNB are also challenged, since QoI resistance has developed in the pathogens that cause both diseases (Blixt et al., 2009; Sierotzki et al., 2007). Therefore, cropping of cultivars with better host resistance, combined with an understanding of the dynamics and the importance of the three leaf blotch diseases within the agronomic environment, is essential for sustainable intensification of wheat production. Indeed, the recent development of molecular tools for quantitative assessment of Pn (Oliver et al., 2008) and Zt (Bearchell et al., 2005) using DNA extracted from infected leaf samples now allows both visual and molecular assessments to be undertaken to efficiently determine the occurrence and nature of mixed field infections. Although a molecular assay for Ptr detection has been published (Antoni et al., 2010), as far as we are aware no validated assay for quantitative Ptr detection is available.
With the aim of testing the hypothesis that mixed necrotrophic fungal infection of European wheat is commonly overlooked by visual assessment alone, the main aims of this study were to: (1) determine the distribution and importance of the three necrotrophic wheat diseases in different environments across Europe; (2) develop a qPCR assay for Ptr; (3) compare visual disease assessments with data from qPCR methods; and (4) assess susceptibility to the three pathogens of a common set of cultivars/breeding lines across the regions studied.
Methods
Leaf sampling
The field survey consisted of wheat flag leaf samples collected from 330 sites over three seasons (2015, 2016 and 2017) (Table 1). Leaf samples were collected at Zadoks growth stage GS73 (early milk) to GS75 (medium milk) from field sites predominantly across four EU countries: Denmark (DK), Norway (NO), Germany (DE) and the United Kingdom (UK). In 2015, samples from Latvia (LV), Finland (FI) and Sweden (SE) were also included. In the first season, sampling in DK and DE targeted fields of high risk of TS, with reduced tillage and wheat as pre-crop. The rest of the samples across all countries were taken from fields representing standard agricultural practices in the regions from both untreated and fungicide-treated fields. Samples were collected from winter wheat varieties, with additional sampling from spring wheat varieties in Norway.
Quantification of disease level
The disease severity of each of the three pathogens was assessed visually on the surveyed flag leaf materials using a percentage rating scale from 0% (no symptoms) to 100% (severe disease). Subsequently, leaves were air dried at room temperature and stored in paper bags for further use. A leaf segment of 8 cm was cut from each leaf, measured from the leaf tip. Segments from 10 leaves per site were pooled for further investigation. Grinding of the leaf material was performed using a GenoGrinder (SPEX SamplePrep, USA) using 10 steel balls (ø 4–5 mm) at 1500 rpm for 2 × 4 min. DNA was extracted from 25 mg of powdered leaf material suspended in 150 μl of lysis buffer using a Sbeadex® Mini Plant Kit (LGC Genomics, Germany) following the manufacturer’s instructions, automated with a KingFisher™ Magnetic Particle Processor (Thermo Fisher Scientific, USA).
Quantification of Pn and Zt was undertaken by qPCR using primers and probes targeting the β-tubulin gene (Bearchell et al., 2005). Primers FDTR321 5’-CGAAGTACGTCAACCGCTTCT-3′, RDTR390 5’-TTGGACGCCTATTGCATGTTAG-3′ and the TaqMan probe DTR345 5’-FAM-CAAGCTCCCGATAAGCGGTCAAT–TAMRA targeting the Ptr β-tubulin gene were designed in this study using GenBank accession JQ314403. This sequence was aligned with β-tubulin sequences of Zt, Pn and other closely related species identified by BLASTn search in GenBank using CLC Main Workbench 8.0 (QIAGEN Bioinformatics), and primers were designed using Primer Express (Applied Biosystems). Primer and probe specificities were tested by BLASTn searches in GenBank and by performing quantitative polymerase chain reaction (qPCR) on a range of Ptr, Pn and Zt isolates. No amplification was seen from non-target DNA (Supplementary Table 1). For normalization to compensate for variation in DNA extraction and qPCR efficiencies, quantification of plant DNA using a qPCR assay targeting the plant EF1α and a standard curve made from wheat DNA was performed as described by Nicolaisen et al. (2009), and the amount of fungal DNA calculated as pg of fungal DNA per ng of plant DNA.
Each qPCR reaction was performed in a total volume of 15 μl consisting of 7.5 μl of TaqMan™ Universal Mastermix (Applied Biosystems), 1.35 μl of forward primer FDTR321 (10 μM), 1.35 μl of reverse primer RDTR390 (10 μM) and 0.75 μl of probe DTR345 (5 μM), 2.05 μl of Milli-Q water, and 2.0 μl of undiluted template DNA. Duplicate reactions were undertaken for each sample to confirm reproducibility, and qPCR was run on an Applied Biosystems ViiA7 using the following program: initial cycle at 50 °C for 2 min; another cycle at 95 °C for 10 min; followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Five-fold dilution series of DNA extracted from pure cultures of Ptr, Pn and Zt were used to determine standard curves for quantification. The DNA concentration of the stock solution was measured with a NanoDrop™ 1000 (Thermo Scientific™). Based on standard curves the amplification efficiency E was 95% (Ptr), 83% (Pn) and 90% (Zt) respectively, and R2 ranged from 0.992 to 0.997.
Molecular screening of isolates for Ptr effectors
Single-spore isolates of Ptr were isolated from leaf samples collected in DE, DK and UK during 2015 and 2016. Leaves were incubated on moist filter paper in petri dishes. Single spores were transferred from lesions to grass agar (GA) prepared as described by Thach et al. (2013) using a sterile needle and incubated at 17 °C (12 h dark light/ 12 h UV light). After approximately 2 weeks, mycelium was scraped off the plate, frozen in liquid N2 and stored at −80 °C. The mycelium was pulverized using a GenoGrinder at 1500 rpm for 30 s. After grinding, the powder was stored in liquid N2 before addition of lysis buffer and DNA extraction as described above for leaf samples. PCR for presence of the ToxA gene in Ptr was performed using the primers ToxAscreeningF and ToxAscreeningR as described by Antoni et al. (2010). The presence of ToxB in Ptr was tested using the primers TB10 and TB12 according to Martinez et al. (2001). All isolates were also tested with the Ptr-specific primers PtrUniqueF2 and PtrUniqueR2 to confirm presence of DNA and species identity (Antoni et al., 2010). Isolates containing ToxA (isolate ‘ToxA’) and ToxB (DW5) were included as positive controls. PCR products were visualized after electrophoresis across a 1.5% agarose gel with SYBR stain.
Screening common cultivars for disease resistance
During the 2016, 2017 and 2018 seasons, 40 winter wheat genotypes were screened in field plots for resistance to TS, STB and SNB in DK, DE, UK and NO (Supplementary Table 2). The genotypes mainly represented wheat cultivars released and widely grown in the four countries. The founder varieties of the multi-parental ‘BMWpop’ (Stadlmeier et al., 2018) and ‘NIAB Elite MAGIC’ (Mackay et al., 2014) populations were also included in the panel. Each cultivar was present in at least two replicate plots at each of the disease resistance screening trials in the four countries, as previously described (Lin et al., 2020; Stadlmeier et al., 2019). Different inoculation methods for the three diseases were applied depending on year and location (Supplementary Table 3). The scoring of disease severity was based on visual assessment of the percentage leaf area covered by necrotic and chlorotic lesions using a rating scale from 0% (no symptoms) to 100% (severe disease). Infection was based on assessment of the flag leaves between Zadoks GS 75–79. Phenotypic data were adjusted in R (R Development Core Team, 2017) for block (randomized complete block and alpha lattice design) and replication effect in each year and country. Disease severity was visualized using the R/ComplexHeatmap package (Gu et al., 2016). The grouping of the genotypes was based on hierarchical clustering using the Euclidean distance matrix and the clustering method ‘average’.
Results
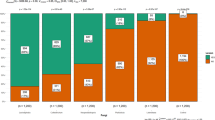
During the field seasons 2015, 2016 and 2017, 330 samples from sites located predominantly in DK, DE, NO and UK (Table 1) were analyzed visually and by qPCR for the assessment of STB, SNB and TS disease severity and infestation rate. Based on qPCR results, one, two, and three pathogens were detected in 31%, 25%, and 41% of all samples, respectively (Table 2). Although 65% of all samples contained more than one of the three species, one of them was usually dominant (Supplementary Table 4). The average visual assessments per disease and country (Figs. 1, 2) correlated relatively well with the amounts of fungal DNA (STB R2 = 0.30, TS R2 = 0.17, SNB R2 = 0.13) (Supplementary Fig. 1). However, for the specific diseases including data from all countries and years, the correlations were only low to moderate; for TS, SNB and STB, R2 = 0.17, 0.13 and 0.32, respectively. This likely reflects the difficulty in visually distinguishing symptoms in mixed infection, particularly TS and SNB. Disease levels were most severe in 2017 for all countries, which is reflected in the higher values detected by qPCR (Fig. 1; Supplementary Table 4).
Box-plots showing the amount of fungal DNA (pg) of Pyrenophora tritici-repentis (Ptr), Zymoseptoria tritici (Zt) and Parastagonospora nodorum (Pn) in Germany (DE, blue), Denmark (DK, green), Norway (NO, orange), and the United Kingdom (UK, purple) in the years 2015, 2016 and 2017. Solid rectangles indicate the 25 and 75 percentiles and the horizontal lines that extend below and above from them indicate 5 and 95 percentiles, respectively. The horizontal lines within the solid rectangles indicate the median. Dots represent outliers. Note the change in scale on the y-axis used for the 2017 season dataset
Analysis of qPCR results found STB to be the most widespread and severe of the three diseases investigated, present in 90% of all samples (Table 3). Although levels were most severe in UK samples, with a maximum of 40% mean disease severity based on visual assessment and presence in 100% of samples based on qPCR, the disease was also highly prevalent in DK, DE and NO (Table 3 and Fig. 1).
On average, qPCR found Pn and Ptr to occur at similar levels, present in 54% and 57% of all samples, respectively (Table 3). In the samples from NO, Pn was very common and most abundant with an almost 100% detection rate in qPCR and a maximum of the mean disease severity of 29% (NO 2017, winter wheat) (Table 3 and Fig. 1). In all other regions, SNB played a minor role with mean disease severity scores found to be between 0% (DK, DE, UK) and 8% (FI, LV and SE), although Pn was detectable in up to 88% (DE 2017) of the samples within a year and country (Fig. 2).
TS was most pronounced in the samples from DE, with mean disease severity of up to 27% (DE 2015), molecular detection rates of ≥93%, and a high mean level of fungal DNA - especially in Norway in 2017, with 106 pg/ng plant DNA. TS was common in Danish leaf samples (positive qPCR samples per year ranged from 62 to 91%) with a prevalence of high values in samples originating from fields with minimal tillage and pre-crop winter wheat (Supplementary Table 4). In Norway, positive qPCR samples for Ptr per year ranged from 50 to 90%. Very low amounts of Ptr DNA were found in all years in the UK samples, and were present in up to 20% of samples from any one year. Disease levels of TS were not available for 2015 and 2016 in the UK. NO samples included both winter and spring wheat, in contrast to the samples from all other countries which were all winter wheat. Zt was less pronounced in spring wheat, while Pn was present at similar levels in both. Ptr occurred at low levels in both winter and spring wheat (Table 3). From 2015, eight samples from SE, FI, and LV were included in the analysis. The samples showed low disease severities of 1–8% molecular presence of all three pathogens (Table 3).
Screening wheat cultivars for disease susceptibility
A common set of 40 winter wheat genotypes was tested at sites within DE, DK, NO and UK over three seasons to assess resistance to the three pathogens (Fig. 3). Across the 20 trials, broadly similar trends in variety disease resistance rankings were observed for the three pathogens and four countries, which represent a range of climatic conditions. Of the six tan spot trials (DK16, DK17, DE16, DE17, UK17, UK18), the most severe disease levels were observed in Denmark, with only a small number of cultivars showing robust levels of resistance, most notably cv. Creator. Infection was lower in the German TS trials than those in Denmark, and some notable differences in the disease resistance rankings for individual varieties were apparent, most notably for Bussard, Tarso, Nakshov and Hereward. TS disease levels in the two UK trials were generally low, even on varieties previously known to be susceptible, and so we assume likely did not fully challenge cultivar resistance. All SNB trials were carried out in Norway and the UK and the resistance rankings were similar across the three seasons studied, with the most severe attack observed in 2017. Varieties with notable resistance were Dichter, Format, Torp and Jenga. In the two German trials, common trends in cultivar STB resistance rankings were observed across both seasons. A subset of 14 lines (Dichter, Format, Bombus, FIRL3565, KWS MAGIC, Creator, Spontan, Hereford, Substance, Alchemy, Ambition, Jensen, Torp and Jenga) was found to show comparatively good resistance for all three diseases across almost all of the trials undertaken, with Creator performing the best overall.
Percentage disease severity of tan spot (TS), septoria tritici blotch (STB), and septoria nodorum blotch (SNB) across the 40 wheat genotypes assessed in Denmark (DK), Germany (DE), the United Kingdom (UK) and Norway (NO) in 2016, 2017 and 2018. The varieties were chosen to represent wheat cultivars released and widely grown in the four target countries. Disease severity is indicated using a color scale from yellow (no symptoms, 0% infection of the leaves) to red (severe disease, 100% infection of the leaves). Grey represents missing data. The dendrogram shows the clustering of the genotypes according to disease severity
Screening P. tritici-repentis isolates for PtrToxA and PtrToxB
To begin to investigate the effector complement in Ptr within the target geographic region, the pathogen was isolated from leaf material sampled in 2015 and 2016 in DK, DE and the UK and screened for presence/absence of PtrToxA and PtrToxB with gene-specific PCR primers (Table 4). Among the resulting 142 isolates, we found PtrToxA to be present in all UK isolates, while the PtrToxA frequency was generally lower in DK (12% and 26%) than in DE (35% and 42%) in 2015 and 2016, respectively. None of the 142 isolates tested contained the PtrToxB gene.
Discussion
Changes in European patterns of STB, SNB and TS prevalence in wheat have been dramatic since the beginning of the 1980s. Before this time, SNB was considered the most important wheat leaf blotch disease in Northern Europe, while STB was most prevalent in Mediterranean climates (Bayles, 1991; Bearchell et al., 2005; Scharen, 1999). Over the last 30–40 years there has been a focal shift in most North Western European countries from SNB to STB (Bearchell et al., 2005; Shaw et al., 2008; Torriani et al., 2015), although Scandinavia has remained a SNB hot spot (Lin et al., 2020). Over the same period, TS has also become increasingly prominent. At the beginning of the 1990s TS started to become a significant problem in parts of Germany, France, Luxembourg, and Belgium (Jarroudi et al., 2013; Maraite et al., 1992; Wolf & Hoffmann, 1993), as well as in eastern European countries such as Latvia, Czech Republic, Hungary Ukraine and Russia (Sarova et al., 2003). In Denmark and Sweden, TS was first recognized as a disease of increasing significance in the mid-1990s, with the first recognized severe attack occurring in 1999 (Jensen et al., 2001) and in UK it has seen increases in incidence from 2005 (Turner, 2008). The reasons for these regional changes in the prevalence of these three diseases are not clearly understood but are attributed, most commonly, to increased use of minimal tillage practices. The qPCR results of our field surveys across DK, DE, NO and UK verified that all three diseases are common, although the prevalence varies between regions, and between years. Disease levels were most severe in 2017 across all tested countries, likely reflecting favorable conditions for the target diseases in this season. STB is today considered as the most yield-reducing disease in winter wheat in northern Europe (Fones & Gurr, 2015; Jørgensen et al., 2014), and this is in line with its dominating role found in this study. In our survey, qPCR indicated that levels of Z. tritici DNA are notably higher in the UK compared with samples from the other countries targeted. With a few exceptions, STB was present in all samples - the levels in DK, DE and NO were quite similar for winter wheat, but very low for spring wheat from NO.
The level of SNB was generally low, except for NO which had a high level of SNB in both winter and spring wheat. The eight samples from SE, FI, and LV also contained relatively high levels of Pn DNA, indicating a similar common appearance of this pathogen. Even though SNB is rarely recognized as a pathogen in DE, DK and UK, the disease has not disappeared and could be detected in 80%, 33% and 7% of the tested samples, respectively. In NO, the pathogen could be detected in 99% of the samples. For reasons which are not clear, SNB has stayed more significant in NO in comparison with most other regions of northern Europe. One notable difference is that more spring wheat is grown than winter wheat in Norway, while winter wheat dominates in the other surveyed countries. Furthermore, the observation that the ToxA sensitivity locus Tsn1 is present at notably higher frequency in Norwegian wheat cultivars (Lin et al., 2020; Ruud et al., 2018) than in wider northern European germplasm (Downie et al., 2018), and that a high proportion of Norwegian P. nodorum populations carry ToxA (>67%, Ruud et al., 2018; Lin et al., 2020), indicates that the ToxA/Tsn1 interaction may be a contributing factor to the predominance of SNB in Norway. It has been suggested that the higher occurrence of SNB relative to STB in spring wheat in Norway is because the comparatively long Z. tritici latent period (Cunfer, 1999) does not allow time for STB to establish as quickly as SNB within the relatively short Norwegian spring wheat growing period (Lin et al., 2020). This fits with the results of our field survey here, where Norwegian STB disease severity based on visual assessment was almost always recorded just in winter wheat. Although the more sensitive qPCR analysis found Z. tritici to also be present in the Norwegian spring wheat samples, it was at lower levels than P. nodorum (Table 3).
We found P. tritici-repentis to be present in 94%, 75% and 71% of the samples from DE, DK and NO, respectively, and to be a serious pathogen in fields where minimal tillage and pre-crop wheat is common. TS is not commonly identified by growers as a pathogen in UK, but it could be detected by qPCR in 9% of UK field samples. Although P. tritici-repentis was detected in many fields, the level of fungal DNA was comparatively low in most cases, except for German sites, as well as Danish fields with a history of minimal tillage and pre-crop wheat. We commonly detected all three pathogens via qPCR, even though they were not recognized visually during the season. Which of the three pathogens emerges as prevalent seems to be driven by regional conditions, where we see SNB dominant in Norway, and TS dominant in Germany and reduced tillage sites in Denmark. The first year of the sampling, fields from Germany and Denmark were targeted to include those with known history of TS, mainly from fields with reduced tillage. However, the level of TS in the two subsequent seasons was also high, indicating that TS plays a major role in Germany, in addition to sites with relatively high readings from both Denmark and Norway. It has previously been suggested that TS antagonizes STB (Sutton & Vyn, 1990), particularly in fields with a high density of wheat residue. It has also been seen in competition trials that P. tritici-repentis out-competes P. nodorum (Adee et al., 1990). Zymoseptoria tritici has a latent period of 3–4 weeks (Shaw, 1999) while P. tritici-repentis has a much shorter latent period of 5–8 days (Riaz et al., 1991), hence it seems likely that under optimal conditions TS will also out-compete STB. However, we found this not to be the case in practice, with Z. tritici DNA more abundant than P. tritici-repentis at almost all sites tested (Fig. 1; Supplementary Table 4). Overall, we propose that farming practice such as tillage methods, choice of spring or winter crops and adaptation to specific climate are the main drivers that determine which of the three diseases becomes dominant when mixed infections are evident.
Several recent studies of ToxA, Tox1 and Tox3 revealed alternative roles for these necrotrophic effectors during infection in helping the pathogen to avoid host recognition and defense (McDonald & Solomon, 2018). A chitin-binding domain in Tox1 allows it to bind to P. nodorum chitin and prevent access to host chitinases from generating chitin monomers that trigger PAMP-triggered immunity (PTI) (Liu et al., 2019). On the other hand, ToxA and Tox3 bind to pathogenesis-related 1 (PR1) proteins (Breen et al., 2016; Lu et al., 2014). Perhaps, interaction of ToxA and Tox3 with PR1 proteins allow the fungus to manipulate and suppress host PTI (McDonald & Solomon, 2018). Manipulation of the host defense by P. nodorum ToxA and Tox1 may increase host susceptibility to other pathogens, therefore resulting in the formation of disease complexes. However, this hypothesis requires further testing.
The correspondence between visual percentage disease severity and qPCR estimation of fungal DNA was quite variable, indicating that necrotrophic fungal infection other than the dominating species is commonly overlooked by visual assessment. The Norwegian spring wheat samples were visually scored as having SNB as the major disease, overlooking the fact that TS and STB were also present at relatively high levels, based on estimation of fungal DNA by qPCR. Similarly, visual assessment of the German and Danish samples overlooked that SNB was present in winter wheat. Collectively, this confirms that visual assessment can be challenging where mixed infection occurs. Visual determination is also made harder the later the samples are collected in the season (i.e. after growth stage 75), as general senescence also makes it difficult to visually detect and distinguish the three diseases. Furthermore, asymptomatic presence of the fungus causing no, or very weak, visual symptoms may also occur, especially in the early phases of infection (Duba et al., 2018).
As the three wheat leaf blotch diseases investigated can be simultaneously present in the host, from a disease management point of view it would be beneficial in some agricultural environments if the cultivars commonly grown possessed broad resistance against all three diseases. The varieties showed a wide range of susceptibility to the three diseases, with the top six ranked cultivars (in terms of highest disease scores) showing a high degree of susceptibility to all three diseases. A higher number of cultivars showed relatively good resistance to both STB and SNB. However only very few cultivars showed resistance to all three diseases, e.g. ‘Creator’.
Finally, while surveys of P. nodorum effectors have now been relatively well studied in northern Europe, little research has been done on the effector complement of P. tritici-repentis. In our survey, none of the 142 isolates tested carried PtrToxB, in line with previously published reports of the absence of ToxB in isolates from Australia (Antoni et al., 2010), New Zealand (Weith, 2015), Romania, Latvia and Lithuania (Abdullah et al., 2017). However, isolates carrying PtrToxB have been reported in various countries, predominantly in the Middle East (Azerbaijan, Turkey [Lamari et al., 2003], Algeria [Lamari et al., 1995, 2003; Benslimane, 2018], Syria, Turkey [Lamari et al., 2003; Lamari, Strelkov, et al., 2005a]) and North America (Canada [Lamari, McCallum, & Depauw, 2005b] and the USA [Ali et al., 1999; Abdullah, 2017]). A recent survey of northern European wheat varieties found ToxB sensitivity to be rare, present in just 7% of varieties (Corsi et al., 2020). Therefore, if isolates carrying PtrToxB do spread into Europe, most European wheat varieties are already likely to be insensitive to this effector. In contrast, we found PtrToxA to be present in our survey samples, including all of the UK isolates tested. Screening for and selection against ToxA-sensitive germplasm in wheat breeding programmes has been successfully applied in Australia, where PtrToxA seems to be ubiquitous (Antoni et al., 2010) and ToxA-sensitive varieties previously occupied the majority of the wheat-growing area in Western Australia (Phan et al., 2020). As a result, average yield increased by about 10% (Vleeshouwers & Oliver, 2014). For growers in the countries investigated here, use of varieties that lack the ToxA sensitivity gene Tsn1 could be a useful precaution against susceptibility to TS and SNB, especially if the presence of the pathogen effector genes PtrToxA and PnToxA were found to be increasing prevalent over time. Furthermore, such lines could be used for pre-emptive breeding strategies against spot blotch. Caused by the related wheat fungal pathogen Bipolaris sorokiniana, spot blotch is currently problematic in Asia and has acquired ToxA via horizontal gene transfer, most likely from P. nodorum (McDonald et al., 2017). However, our finding that only 17% of the Danish isolates and 40% of the German isolates contained PtrToxA, combined with relatively low frequencies of ToxA-sensitive alleles at the Tsn1 locus in most European wheats (~10%, Downie et al., 2018) highlights that screening for cultivar resistance cannot solely focus on the screening of the known P. tritici-repentis effectors (See et al., 2019). The best results for the management of TS, SNB and STB are most likely to arise through integrated programmes, such as the model recently suggested by Downie et al. (2020): (1) Establish contemporary isolate collections and disease outbreak monitoring programmes; (2) use these collections to test for cultivar resistance and establish the presence of new effectors; (3) monitor for changes in pathogen populations using high-throughput genotyping methodologies; (4) grow wheat varieties with differing genetic backgrounds to avoid buildup of a specialized pathogen population, especially where minimum tillage is common; (5) where local pathogen populations contain known effector genes, grow wheat varieties that carry insensitive alleles at the corresponding host loci; and (6) continue wheat research and development activities to exploit additional sources of host genetic resistance.
Overall, our study highlights that mixed infection of wheat with two or more of the fungal pathogens causing the three blotch diseases investigated is common, despite being overlooked by visual inspection. This finding clearly has implications for the future management of wheat diseases and may also apply to other crops. In the assessment of crop varieties for disease resistance and efficacy trials of fungicides and other crop-protection measures it should be taken into account that visually assessed symptoms may not represent the entirety of the causal organisms. Molecular techniques to measure fungal biomass of multiple species simultaneously may need to be used in routine settings. These observations also raise interesting questions about the interactions of these diseases. Do the various pathogens compete when co-infections occur or is there synergy? Is there suppression by the dominant pathogen of sporulation of the other pathogens? Does sporulation of different pathogens occur at different times and/or places (sporulation suppression with/without refugia)? Or is the sporulation of the weaker pathogen simply overlooked by pathologists? The answers to these questions will surely be needed to help establish improved breeding and management practices for regions where mixed leaf blotch infection is common.
References
Abdullah, S. (2017). Characterization of Pyrenophora tritici-repentis in wheat and rye to study tan spot susceptibility and insights into its relationship with stem rust resistance. Theses and Dissertations, 1183.
Abdullah, S., Sehgal, S. K., Ali, S., Liatukas, Z., Ittu, M., & Kaur, N. (2017). Characterization of Pyrenophora tritici-repentis (tan spot of wheat) races in Baltic states and Romania. Plant Pathology Journal, 33, 133–139.
Abdullah, A. S., Turo, C., Moffat, C. S., Lopez-Ruiz, F. J., Gibberd, M. R., Hamblin, J., & Zerihun, A. (2018). Real-time PCR for diagnosing and quantifying co-infection by two globally distributed fungal pathogens of wheat. Frontiers in Plant Science, 9, 1–10.
Adee, S. R., Phender, W. F., & Harnett, D. C. (1990). Competition between Pyrenophora tritici-repentis and Septoria nodorum in the wheat leaf as measured with de Wit replacement series. Phytopathology, 80, 1177–1182.
Ali, S., Francl, L. J., & Dewolf, E. D. (1999). First report of Pyrenophora tritici-repentis race 5 from North America. Plant Disease, 83, 591.
Antoni, E. A., Rybak, K., Tucker, M. P., Hane, J. K., Solomon, P. S., Drenth, A., Shankar, M., & Oliver, R. P. (2010). Ubiquity of ToxA and absence of ToxB in Australian populations of Pyrenophora tritici-repentis. Australasian Plant Pathology, 39, 63–68.
Bayles, R. A. (1991). Varietal resistance as a factor contributing to the increased importance of Septoria tritici Rob. and Desm. In the UK wheat crop. Plant Varieties and Seeds, 4, 177–183.
Bearchell, S. J., Fraaije, B. A., Shaw, M. W., & Fitt, B. D. L. (2005). Wheat archive links long-term fungal pathogen population dynamics to air pollution. Proceedings of the National Academy of Sciences U S A, 102, 5438–5442.
Benslimane, H. (2018). Virulence phenotyping and molecular char-acterisation of a new virulence type of Pyrenophora tritici-repentis the causal agent of Tan spot. Plant Pathology Journal, 34, 139–142.
Blixt, E., Djurle, A., Yuen, J., & Olson, Å. (2009). Fungicide sensitivity in Swedish isolates of Phaeosphaeria nodorum. Plant Pathology, 58, 655–664.
Blixt, E., Olson, Å., Lindahl, B., Djurle, A., & Yuen, J. (2010). Spatiotemporal variation in the fungal community associated with wheat leaves showig sympotms similar to stagonospora nodorum blotch. European Journal of Plant Pathology, 126, 373–386.
Breen, S., Williams, S. J., Winterberg, B., Kobe, B., & Solomon, P. S. (2016). Wheat PR-1 proteins are targeted by necrotrophic pathogen effector proteins. The Plant Journal, 88, 13–25.
Brown, J. K., Chartrain, L., Lasserre-Zuber, P., & Saintenac, C. (2015). Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genetics and Biology, 79, 33–41.
Cools, H., & Fraaije, B. A. (2013). Update on mechanisms of azole resistance in Mycosphaerella graminicola and implications for future control. Pest Management Science, 69, 150–155.
Corsi, B., Percival-Alwyn, L., Downie, R. C., Venturini, L., Iagallo, E. M., Campos Mantello, C., McCormick-Barnes, C., See, P. T., Oliver, R. P., Moffat, C. S., & Cockram, J. (2020). Genetic analysis of wheat sensitivity to the ToxB fungal effector from Pyrenophora tritici-repentis, the causal agent of tan spot. Theoretical and Applied Genetics, 133, 935–950.
Cunfer BM (1999). Stagonospora and septoria pathogens of cereals: The infection process. In: Septoria and Stagonospora Diseases of Cereals: A Compilation of Global Research, eds M. van Ginkel, A. McNab, and J. Krupinsky (Mexico: CIMMYT), 41–45.
De Wolf, E. D., Effertz, R. J., Ali, S., & Francl, L. J. (1988). Vistas of tan spot research. Canadian Journal of Plant Pathology, 20, 349–370.
Downie, R. C., Bouvet, L., Furuki, E., Gosman, N., Gardner, K. A., Mackay, I. J., Mantello, C. C., Mellers, G., Phan, H. T. T., Rose, G. A., Tan, K.-C., Oliver, R. P., & Cockram, J. (2018). Assessing European wheat sensitivities to Parastagonospora nodorum nectrophic effectors and fine-mapping the Snn3-B1 locus conferring sensitivity to the effector SnTox3. Frontiers in Plant Science, 9, 881.
Downie, R. C., Lin, M., Corsi, B., Ficke, A., Lillemo, M., Oliver, R. P., Phan, H., Tan, K.-C., & Cockram, J. (2020). Spetoria nodorum blotch of wheat: Disease management and resistance breeding in the face of shifting disease dynamics and a changing environment. Phytopathology. https://doi.org/10.1094/PHYTO-07-20-0280-RVW.
Duba, A., Goriewa-Duba, K., & Wachowska, U. (2018). A review of the interaction between wheat and wheat pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum. International Journal of Molecular Sciences, 19, 1138.
Faris, J., Liu, Z., & Xu, S. (2013). Genetics of tan spot resistance in wheat. Theoretical and Applied Genetics, 126, 2197–2217.
Fones, H., & Gurr, S. (2015). The impact of Septoria tritici blotch disease on wheat: An EU perspective. Fungal Genetics and Biology, 79, 3–7.
Fraaije, B. A., Cools, H. J., Fountaine, J., Lovell, D. J., Motteram, J., West, J. S., & Lucas, J. A. (2005). Role of ascospores in further spread of QoI-resistant cytochrome b alleles (G143A) in field populations of Mycosphaerella graminicola. Phytopathology, 95, 933–941.
Friesen, L. F., Meinhardt, S. W., & Faris, J. H. (2007). The Stagonospora nodorum-wheat pathosystem involves multiple proteinaceous host-selective toxins and corresponding host sensitivity genes that interact in an inverse gene-for-gene manner. The Plant Journal, 51, 681–692.
Gu, Z., Eils, R., & Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics, 32, 2847–2849.
HGCA (2021) Recommended List: https://ahdb.org.uk/rl
Jarroudi, M., Kouadio, L., Beyer, M., Geraud, F., Tychon, B., & Deelefoss, P. (2013). Factors affecting Tan spot on winter wheat in the grand-duchy of Luxembourg. Journal of Plant Pathology, S1, 7–17.
Jensen, K. F., Jørgensen, L. N., Henriksen, L., & Nielsen, G. C. (2001). Hvedebladplet—En ny svampesygdom i Danmark. Grøn Viden, Markbrug nr., 232 (In Danish).
Jørgensen, L. N., & Olsen, L. V. (2007). Control of tan spot (Drechslera tritici-repentis) using host resistance, tillage methods and fungicides. Crop Protection, 26, 1606–1616.
Jørgensen, L. N., Hovmøller, M. S., Hansen, J. G., Lassen, P., Clark, B., Bayles, R., Rodemann, B., Jahn, M., Flath, K., Goral, T., Czembor, J., Cheyron, P., Maumene, C., Pope, C., Nielsen, G. C., & Berg, G. (2014). IPM strategies and their dilemmas including an introduction to www.eurowheat.org. Journal of Integrative Agriculture, 13, 265–281.
Kollers, S., Rodemann, B., Ling, J., Korzun, V., Ebmeyer, E., Argillier, O., Hinze, M., Plieske, J., Kulosa, D., Ganal, M. W., & Röder, M. S. (2013). Genetic architecture of resistance to Septoria tritici blotch (Mycospherella graminicola) in European winter wheat. Molecular Breeding, 32, 411–423.
Krupinsky, J. M., & Berdahl, J. D. (1984). Evaluation of Agropyron intermedium for reactions to various leaf spot diseases. Plant Disease, 68, 1089–1091.
Lamari, L., Sayoud, R., Boulif, M., & Bernier, C. C. (1995). Identification of a new race in Pyrenophora tritici-repentis: Implications for the current pathotype classification system. Canadian Journal of Plant Pathology, 17, 312–318.
Lamari, L., Strelkov, S., Yahyaoui, J., Orabi, J., & Smith, R. B. (2003). The identification of two new races of Pyrenophora tritici-repentis from the host center of diversity confirms a one-to-one relation-ship to tan spot of wheat. Phytopathology, 93, 391–396.
Lamari, L., Strelkov, S. E., Yahyaoui, A., Amedov, M., Saidov, M., Dju-nusova, M., & Koichibayev, M. (2005a). Virulence of Pyrenophora tritici-repentis in the countries of the silk road. Canadian Journal of Plant Pathology, 27, 383–388.
Lamari, L., McCallum, B. D., & Depauw, R. M. (2005b). Forensic pathology of Canadian bread wheat: The case of tan spot. Phytopathology, 95, 144–152.
Lin, M., Ficke, A., Cockram, J., & Lillemo, M. (2020). Genetic structure of the Norwegian Parastagonospora nodorum populations. Frontiers in Microbiology, 11. https://doi.org/10.3389/fmicb.2020.01280.
Liu, Z., Gao, Y., Kim, Y. M., Faris, J. D., Shelver, W. L., de Witt, P. J. G. M., Xu, S. S., & Friesen, T. L. (2019). SnTox1, a Parastagonospora nodorum necrotrophic effector, is a dual-function protein that facilitates infection while protecting from wheat-produced chitinases. New Phytologist, 211, 1052–1064.
Loughman, R., Wilson, R. E., & Thomas, G. J. (1994). The influence of disease complexes involving Leptosphaeria (Septoria) nodorum on detection of resistance to three leaf spot diseases in wheat. Euphytica, 72, 31–42.
Lu, S., Faris, J. D., Sherwood, R., Friesen, T. L., & Edwards, M. C. (2014). A dimeric PR −1-type pathogenesis-related protein interacts with ToxA and potentially mediates ToxA -induced necrosis in sensitive wheat. Molecular Plant Pathology, 15, 650–663.
Mackay, I. J., Bansept-Basler, P., Barber, T., Bentley, A. R., Cockram, J., Gosman, N., et al. (2014). An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: Creation, properties, and validation. G3 Genes|Genomes|Genetics, 4, 1603–1610.
Maraite, H., Berny, J. F., & Goffin, A. (1992). Epidemiology of tan spot in Belgium. In L. J. Franci, J. M. Krupinsky, & M. P. McMullen (Eds.), Advances in Tan spot research, Proceedings of the second international Tan spot workshop, June 25–26 (Vol. 1992, pp. 73–79). North Dakota State University.
Martinez, J. P., Ottum, S. A., Ali, S., Franci, L. J., & Ciuffetti, L. M. (2001). Characterization of the ToxB gene from Pyrenophora tritici-repentis. Molecular Plant Microbe Interactions, 14, 675–677.
McDonald, M. C., & Solomon, P. S. (2018). Just the surface: Advances in the discovery and characterization of necrotrophic wheat effectors. Current Opinions in Microbiology, 46, 14–18.
McDonald, M. C., Ahren, D., Simpfendorfer, S., Milgate, A., & Solomon, P. S. (2017). The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Molecular Plant Pathology, 19, 432–439.
Moreno MV, Stenglein SA, Perelló AE (2012). Pyrenophora tritici-repentis, causal agent of Tan spot: A review of intraspecific genetic diversity. The molecular basis of plant genetic diversity, prof. Mahmut. Caliskan (Ed.), ISBN: 978-953-51-0157-4, InTech, Available from: http://www.intechopen.com/books/themolecular-basis-of-plant-genetic-diversity/pyrenophora-tritici-repentis-causal-agent-of-tan-spot-a-review-ofintraspecific-genetic-diversity.
Nicolaisen, M., Supronienė, S., Nielsen, L. K., Lazzaro, I., Spliid, N. H., & Justesen, A. F. (2009). Real-time PCR for quantification of eleven individual Fusarium species in cereals. Journal of Microbiological Methods, 76, 234–240.
Oliver, R. P., Rybak, K., Shankar, M., Loughman, R., Harry, N., & Solomon, P. S. (2008). Quantitative disease resistance assessment by real-time PCR using the Stagonospora nodorum-wheat pathosystem as a model. Plant Pathology, 57, 527–532.
Phan, H. T. T., Jones, D. A. B., Rybak, K., Dodhia, K. N., Lopez-Ruiz, F. J., Valade, R., Gout, L., Lebrun, M.-H., Brunner, P. C., Oliver, R. P., & Tan, K.-C. (2020). Low amptitude boom-and-bust cycles define the septoria nodorum blotch interaction. Frontiers in Plant Science, 10, 1785.
R Development Core Team. (2017). R: A language and environment for statistical computing. R foundation for statistical computing.
Rehfus, A., Strobel, D., Bryson, R., & Stammler, G. (2017). Mutations in sdh genes in field isolates of Zymoseptoria tritici and impact on the sensitivity to various succinate dehydrogenase inhibitors. Plant Pathology, 67, 175–180.
Riaz, M., Bockus, W. W., & Davis, M. A. (1991). Effects of wheat genotype, time after inoculation and leaf age on conidia production of Drechslera tritici-repentis. Phytopathology, 81, 1298–1301.
Ruud, A. K., Windju, S., Belova, T., Friesen, T. L., & Lillemo, M. (2017). Mapping of SnTox3-Snn3 as a major determinant of field susceptibility to Septoria nodorum leaf blotch in the SHA3/CBRD x Naxos population. Theoretical and Applied Genetics, 130, 1361–1374.
Ruud, A. K., Dieseth, J. A., & Lillemo, M. (2018). Effects of three Parastagonospora nodorum necrotrophic effectors on spring wheat under norwegian field conditions. Crop Science, 58, 159–168.
Saintenac, C., Lee, W. S., Cambon, F., Rudd, J. J., King, R. C., Marande, W., Powers, S. J., Bergès, H., Phillips, A. L., Uauy, C., Hammond-Kosack, K. E., Langin, T., & Kanyuka, K. (2018). Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nature Genetics, 50, 368–374.
Sapkota, R., Knorr, K., Jørgensen, L. N., O’Hanlon, K. A., & Nicolaisen, M. (2015). Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytologist, 207, 1134–1144.
Sarova, J., Hanzalova, A., & Bartos, P. (2003). Incidence of wheat leaf spot pathogens in the Czech Republic. Cereal Research Communications, 31, 145–151.
Scharen AL (1999). Biology of the Septoria/Stagonospora pathogens: an overview. Proceedings of 5th International Septoria workshop. CIMMYT, Mexico,19–22.
See, P. T., Iagallo, E. M., Oliver, R. P., & Moffat, C. S. (2019). Heterologous expression of the Pyrenophora tritici-repentis effector proteins ToxA and ToxB, and the prevalence of effector sensitivity in Australian cereal crops. Frontiers in Microbiology, 10, 182.
Serfling A, Kopahnke D, Habekuss A, Novakazi F Ordon F (2017). Wheat diseases: An overview. In: Achieving sustainable cultivation of wheat volume 1. : P. Langridge. Burleigh Dodds Science Publishing, 319–326.
Shaw MW (1999). Population dynamics of Septoria in the crop ecosystem. In: Lucas JA, Bowyer P and Anderson HM (eds) Septoria on cereals: A study of Pathosystems (pp 82–95) CABI Publishing.
Shaw, M. W., Bearchell, S. J., Fitt, B. D. L., & Fraaije, B. A. (2008). Long-term relationships between environment and abundance in wheat of Phaeosphaeria nodorum and Mycosphaerella graminicola. New Phytologist, 177, 229–238.
Sierotzki, H., Frey, R., Wullschleger, J., Palermo, S., Karlin, S., Godwin, J., & Gisi, U. (2007). Cytochrome b gene sequence and structure of Pyrenophora teres and P. tritici-repentis and implications for QoI resistance. Pest Management Science, 63, 225–233.
Singh, R., Singh, P. K., Rutkoski, J., Dp, H., He, X., Jørgensen, L. N., Hovmøller, M. S., & Huerta-Espino, J. (2016). Disease impact on wheat yield potential and prospects of genetic control. Annual Review of Phytopathology, 54, 303–322.
Stadlmeier, M., Hartl, L., & Mohler, V. (2018). Usefulness of a multiparent advanced generation intercross population with a greatly reduced mating design for genetic studies in winter wheat. Frontiers in Plant Science, 9, 1825.
Stadlmeier, M., Jørgensen, L. N., Cors, B., Cockram, J., Hartl, L., & Mohler, V. (2019). Genetic dissection of resistance to the three fungal plant pathogens Blumeria graminis, Zymoseptoria tritici, and Pyrenophora tritici-repentis using a multiparental winter wheat population. G3 Genes|Genomes|Genetics, 5, 1745–1757.
Sutton, J. C., & Vyn, T. J. (1990). Crop sequences and tillage practices in relation to disease of winter wheat in Ontario. Canadian Journal of Plant Pathology, 12, 358–368.
Thach, T., Munk, L., Hansen, A. L., & Jørgensen, L. N. (2013). Disease variation and chemical control of Ramularia leaf spot in sugar beet. Crop Protection, 51, 68–76.
Torriani, S. F. F., Brunner, P. C., McDonald, B. A., & Sierotzki, H. (2009). QoI resistance emerged independently at least 4 times in European populations of Mycosphaerella graminicola. Pest Management Science, 65, 155–162.
Torriani, S. F., Melichar, J. P., Mills, C., Pain, N., Sierotzki, H., & Courbot, M. (2015). Zymoseptoria tritici: A major threat to wheat production, integrated approaches to control. Fungal Genetics and Biology, 79, 8–12.
Turner, J. A. (2008). Proceedings HGCA R&D conference: Arable cropping in a changing climate, 2008.
Vleeshouwers, V., & Oliver, R. (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Molecular Plant-Microbe Interactions, 27, 196–206.
Weith K (2015). Pyrenophora tritici-repentis the causal agent of tan spot: Characterisation of New Zeland populations. . Lincoln University, United Kingdom.
West, J. S., Townsend, J. A., Stevens, M., & Fitt, B. D. L. (2012). Comparative biology of different plant pathogens to estimate effects of climate change on crop diseases in Europe. European Journal of Plant Pathology, 133, 315–331.
Wolf, P. F. J., & Hoffmann, G. M. (1993). Biological studies on Drechslera tritici-repentis (DIED) Shoem (Telemorph Pyrenophora tritici-repentis) (Died) Drechsler, the causal agent of a leaf spot disease of wheat. Zeitschrift für Planzenkrankheiten und Pflanzenschutz, 100, 33–48.
Zhong, Z., Marcel, T. C., Hartmann, F. E., Ma, X., Plissonneau, C., Zala, M., Ducasse, A., Confais, J., Compain, J., Lapalu, N., Amselem, J., McDonald, B. A., Croll, D., & Palma-Guerrero, J. (2017). A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytologist, 214, 619–631.
Acknowledgements
Results were achieved within the ‘EfectaWheat’ project, funded within the framework of the 2nd ERA-CAPs call for Expanding the European Research Area in Molecular Plant Sciences II, with funding from the Biotechnology and Biological Sciences Research Council (BBSRC, grant BB/N00518X/1 to JC), the Research Council of Norway (NFR grant 251894 to ML), the Danish Council of Strategic Research (case number 5147-00002B to LNJ) and the Deutsche Forschungsgemeinschaft (DFG, GZ: HA 5798/2-1, AOBJ: 619206 to LH). Curtin University, Australia, provided the project with in-kind support. Joint coordination and planning of project aims and activities by JC and RO was aided by networking activities funded under the COST Action “SUSTAIN”. Thies Marten Heick kindly provided DNA of some Zymoseptoria tritici isolates.
Author information
Authors and Affiliations
Contributions
JC, LNJ, ML, RO and LH won project funding and designed research. LNJ, ML, VM and MS undertook field surveys. AFJ undertook all qPCR analyses and developed the Ptr qPCR assay. BC, ML and MS undertook disease field trials. BC, JC, AFJ, LNJ and MS analyzed data. AF, LH, SH, IM, JT, K-CT and VM provided project resources and helped guide work. AFJ, LNJ and JC wrote the manuscript with additional inputs from RO. All authors edited and approved the manuscript.
Corresponding authors
Ethics declarations
The authors declare no conflicts of interest and that the research complies with ethical standards. This research does not involve human participants and/or animals.
Supplementary Information
ESM 1
(PDF 383 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Justesen, A.F., Corsi, B., Ficke, A. et al. Hidden in plain sight: a molecular field survey of three wheat leaf blotch fungal diseases in North-Western Europe shows co-infection is widespread. Eur J Plant Pathol 160, 949–962 (2021). https://doi.org/10.1007/s10658-021-02298-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-021-02298-5