The Abundance of Genes Encoding ESBL, pAmpC and Non-β-Lactam Resistance in Multidrug-Resistant Enterobacteriaceae Recovered From Wastewater Effluents
- 1SAMRC Microbial Water Quality Monitoring Centre, University of Fort Hare, Alice, South Africa
- 2Applied and Environmental Microbiology Research Group, Department of Biochemistry and Microbiology, University of Fort Hare, Alice, South Africa
The aquatic environments play a critical function in the widespread of antibiotic-resistant bacteria, ultimately impacting human health. We evaluated the abundance of Enterobacteriaceae and the various resistance gene determinants in final effluents of wastewater treatment plants (WWTPs) in the Eastern Cape Province, Republic of South Africa. A total of 44 presumptive Enterobacteriaceae was recovered following standard isolation methods. Upon molecular confirmation through Matrix-Assisted Laser Desorption/Ionization Time of flight (MALDI TOF), a high predominance of Klebsiella pneumoniae (24%) was noted. All the confirmed isolates (n = 29) subjected to a panel of eighteen antibiotics categorized into eleven different classes were multidrug-resistant (MDR). They displayed resistance against antibiotics in more than three different classes. Notably, one E. cloacae exhibited resistance against all the antibiotics assayed. The multiple antibiotic resistance indices (MARI) ranged from 0.22 to 1.0, indicating an environment with high pressure of antibiotics. Conventional Polymerase Chain Reaction (PCR) analysis showed that 72.4% of the isolates harboured at least one β-lactamase genetic determinant. The most predominant extended-spectrum β-lactamases were blaCTX-M-group (21.5%), blaTEM (20%), and blaSHV (16.9%) while the most predominant plasmid-mediated AmpC-type β-lactamases were blaCIT and blaACC (25.9%) and blaEBC (22.2%). A total of 86.2% of the Enterobacteriaceae harboured a minimum of one non-β-lactam resistance gene determinant with predominance observed in catII (25%), sulII (15.8%), and sulI (14.5%). These results demonstrate an abundance of multidrug-resistant (MDR) Enterobacteriaceae in WWTPs’ effluents. The study confirms the need to optimize current WWTPs’ processes to improve the quality of effluents and ultimately reduce the potential risks of using such water when discharged into the environment.
Introduction
The most common zoonotic pathogens are found in the Enterobacteriaceae family. They are common in water environments and can cause infections in both humans and animals. They can cause various illnesses like diarrhoea, septicaemia, enteritis, and urinary tract infections (Tärnberg, 2012). A typical aquatic environment that harbours these Enterobacteriaceae is the wastewater treatment plant (WWTP), which offers beneficial circumstances for the rapid growth of a diverse bacterial community. These WWTPs are recipients of various categories of wastes from hospitals, households, and industries. Hence, they serve as an essential reservoir of microorganisms that reach the environment through the effluents and infect humans and animals if not adequately treated. It has been reported that WWTPs, even when functioning at optimum, do not eradicate antibiotic-resistant bacteria (ARB) and their genes. Municipal WWTPs have been principally designed to remove nutrients and reduce bacterial load to certain acceptable limits (Okoh et al., 2010; Yu et al., 2018; Aristizábal-Hoyos et al., 2019). When antibiotics are used, the body does not fully metabolize them and are discharged as wastes into the sewer system and finally emptied into the WWTPs, where they are only partially removed (Lamprecht et al., 2014; Adefisoye and Okoh 2016). As a result, antibiotics in wastewater can exert selective pressure on ARB, encouraging their spread in widely varied environments.
Antibiotics of various categories are used to treat infections mediated by the Enterobacteriaceae family. Due to the usage and overuse of antibiotics, ARB proliferation in clinical and veterinary settings arose because of selective pressure generated. The ARB in Enterobacteriaceae is currently considered a significant public health issue with critical priority by the World Health Organization (WHO) (WHO, 2014). There are varying defence mechanisms that bacterial species employ to evade the effects of antibiotics administered against them (Jacoby, 2009). In clinical practice, β-lactam antibiotics are among the most important therapeutic options against infections mediated by members of Enterobacteriaceae (Li et al., 2019). Over time, these microorganisms developed the ability to hydrolyze the chemical backbone in the β-lactams through enzymes, β-lactamases (Rawat and Nair, 2010; Cantón et al., 2012).
β-lactamases are greatly diversified due to their ability to continuously mutate. In this category, the extended-spectrum β-lactamases (ESBLs) have been reported as a great concern in clinicial practice (Jacoby, 2009; Bush and Jacoby, 2010; Cantón et al., 2012). The ESBLs are the class A β-lactamases that can hydrolyze penicillins, penicillin inhibitors, monobactams, and third and fourth-generation cephalosporins and are suppressed by clavulanic acid (Paterson and Bonomo, 2005; Bush and Jacoby, 2010). There has been a significant evolution of these ESBLs with high dissemination potential, especially in clinical settings. The genotype first reported was the TEM (Datta and Kontomichalou, 1965; Bush and Jacoby, 2010), followed by the SHV (Brunton et al., 1986; Bush and Jacoby, 2010). In 1988, the first CTX-M group, the CTX-M-1, was detected (Bauernfeind et al., 1990; Bush and Jacoby, 2010). The CTX-M family, which includes CTX-M-25, CTX-M-9, CTX-M-8, CTX-M-1, and CTX-M-2, is the most predominant ESBL genotype. Several other β-lactamases of clinical importance have been identified. These include the minor ESBLs, which are the PER, GES, and VEB. Other important β-lactamases, such as the carbapenemases, include IMP, KPC, VIM, OXA-1-like, and OXA-48-like (Paterson and Bonomo, 2005; Dallenne et al., 2010). Moreover, the pAmpCs are the class C β-lactamases and include six families; CIT, ACC, EBC, DHA, MOX, and FOX. However, they are closely related to ESBLs but display some differences. In contrast to the ESBLs, the pAmpCs are resistant to a broader range of β-lactams and cannot be inhibited by clavulanic acid (Jacoby, 2009). The β-lactams hydrolyzed by ESBLs, carbapenemases, and pAmpCs are critically important therapeutic agents for humans (WHO, 2017).
The resistance of β-lactamases producers against multiple antibiotic classes makes them of great concern in clinical settings (Rawat and Nair, 2010; Paterson, 2000). It makes the treatment of patients infected by them difficult and often almost impossible. The resistance genes for the ESBLs and pAmpCs are regularly encoded on plasmids with large sizes. These plasmids concurrently encode other resistance genes which belong to other antimicrobials classes. The non-β-lactams include tetracycline, trimethoprim, sulphonamide, chloramphenicol, fluoroquinolone, and aminoglycoside (Kotlarska et al., 2015). The spread of bacterial pathogens concurrently harbouring these resistance genetic determinants is of particular concern as it further limits clinical treatment options. Despite resistance exhibited by bacteria against antibiotics, they still serve as a gold-therapy choice for treating infections caused by bacteria. Therefore, due diligence must be taken to decrease the global dissemination of antibiotic resistance. Therefore, this research aimed to assess the total quantification of the Enterobacteriaceae community present in the WWTPs’ effluents. In addition, to determine the abundance of the ESBLs, pAmpCs, and other non-β-lactam encoding genes in the Enterobacteriaceae isolated at the point of discharge into communal surface water.
Materials and Methods
Description of Study Sites
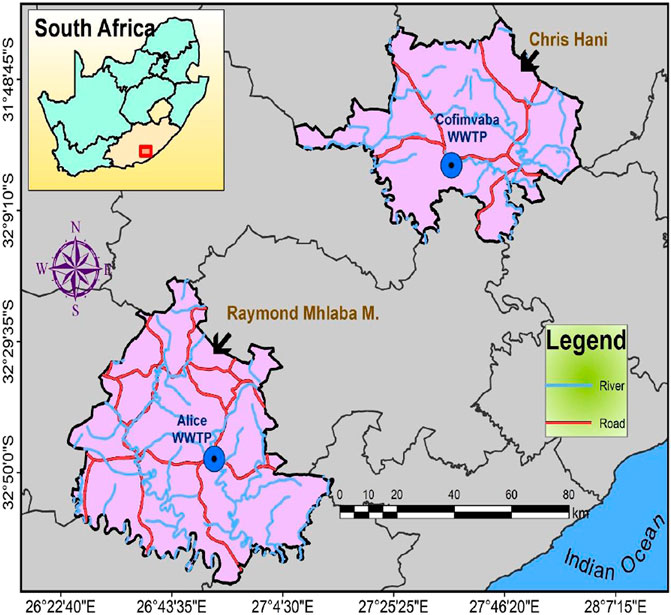
The study was carried out in two District Municipalities (DMs) within the Eastern Cape Province (ECP) Republic of South Africa (RSA), the Amathole and Chris Hani DMs (ADM and CHDM, respectively). In ADM, the WWTP is in Alice, a semi-urban settlement in the Raymond Mhlaba Local Municipality with geographical coordinates 32°50ʹ36ʺ S and 26°55ʹ00ʺ E. The WWTP, which uses the activated sludge technology, receives light industrial wastewater, run-off water, and residential sewage. The WWTP is comprised of a screen channel, aerobic and anaerobic chambers, and a clarifier. The effluent is chlorinated before it is released into the Tyhume River. The plant’s capacity was designed for 2000 m3/day but was operating at about 1,100 m3/day, which is 55% of its designed capacity during the period of sample collection.
The WWTP for the second DM, the CHDM, is located in Cofimvaba, another semi-urban settlement located at 32°03ʹ23ʺ S and 27°60ʹ08ʺ E. Domestic sewage, run-off waters, and some light industrial wastewater are all treated at this facility. It also uses an activated sludge technology treatment facility which includes systems similar to those used at the Alice WWTP described above. The final chlorinated effluent is drained down the Cofimvaba River, which finally empties into the Great Kei River. A map showing the location of the sampling sites is shown in Figure 1.
Sample Collection
A total of six wastewater samples were obtained from WWTPs in October and November 2017. For de-chlorination, the treated final effluents exiting the end-of-pipe were collected into sterile 1-L polypropylene containers into which 1.7 ml of 1 per cent sodium thiosulfate solution has been dispensed (Standard Methods, 2005). Within 6 h of sample collection, they were transported to the lab on ice for microbiological testing. Samples were held at 4°C until preliminary steps were executed.
Quantification and Isolation of Presumptive Enterobacteriaceae
A ten-fold serial dilution was carried out using sterile distilled water. Every 100 ml of adequately diluted samples was filtered using a vacuum filtration pump. The membrane filters were aseptically transferred to the Violet Red Bile Glucose (VRBG) agar (Merck, Modderfontein, RSA) and incubated at 37°C for 18 h. Purple-red colonies were then counted as Enterobacteriaceae, with the results designated as colony-forming units per 100 ml (CFU/100 ml). For the isolation of presumptive Enterobacter spp., Citrobacter spp., E. coli, and Klebsiella spp., the different colonies on VRBG were then picked unto MacConkey agar and Eosin Methylene Blue agar (Conda, Pronadisa, RSA). The purified presumptive isolates were stored at −80°C in 25% glycerol stock for further laboratory analysis.
Molecular Confirmation of Presumptive Enterobacteriaceae
The molecular identities of the purified presumptive isolates were done using Matrix-Assisted Laser Desorption Ionization (MALDI-TOF) coupled with Mass Spectrometry (MS) using the Biotyper 3.0 protocol (Bruker Daltonics, Germany) as previously described (Standing et al., 2013). All isolates were tested in duplicates. For quality control, Bacterial Test Standard (BTS) (8255343) was included in every plate. The manufacturer’s guideline was used in the interpretation of the results obtained. Score values lower than 1.70 were excluded, as the identification is considered unreliable (Standing et al., 2013).
Antimicrobial Susceptibility Profile Using Disk Diffusion Assay
The disk diffusion protocol, following the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2018), was used to determine the antimicrobial susceptibility profile of all the confirmed bacterial isolates after identification using MALDI-TOF MS. The list of antibiotics evaluated were: cefuroxime (CXM/30 μg) and cefotaxime (CTX/30 μg) belonging to the cephems, chloramphenicol (C/30 μg) belonging to the phenicols, amoxicillin/clavulanic acid (AUG/30 μg) and ampicillin (AP/10 μg) belonging to the β-lactams, ciprofloxacin (CIP/5 μg) and norfloxacin (NOR/30 μg) belonging to the fluoroquinolones, doxycycline (DXT/30 μg) and tetracycline (T/30 μg) belonging to the tetracyclines, amikacin (AK/30 μg) and gentamicin (GM/10 μg) belonging to aminoglycosides, colistin (CO/25 μg) and polymyxin B (PB/300 units) belonging to the polymyxins, nitrofurantoin (NI/300 μg) belonging to the nitrofurans, nalidixic acid (NA/30 μg) belonging to the quinolones, meropenem (MEM/10 μg) and imipenem (IMI/10 μg) belonging to the carbapenems and lastly, trimethoprim-sulfamethoxazole (TS/25 μg) belonging to the sulphonamides (Mast Diagnostics, United Kingdom).
Assessment of Multiple Antibiotic Resistance Phenotypes and Multiple Antibiotic Resistance Indices
By analyzing the antibiotic susceptibility of these isolates, an isolate was classified as multidrug-resistant (MDR) when it exhibited resistance against at least one antimicrobial agent in at least three different antibiotic groups, as previously described (Magiorakos et al., 2012). The MARPs were consequently assessed as earlier described (Ateba and Bezuidenhout, 2008). The MARI of the bacterial species was derived as defined following the mathematical calculation (Krumperman, 1983):
Here “c” refers to the sum of antibiotics the bacterial species displayed phenotypic resistance against, while “d” refers to the total number of antibiotics assessed. A MARI score of more than 0.2 indicates that antibiotics are intensively used in that area and implies an environment with a high risk of antimicrobial resistance proliferation (Krumperman, 1983; Osundiya et al., 2013).
DNA Extraction
Genomic DNA used for Polymerase Chain Reaction (PCR) experiment was extracted using the boiling method as reported previously (Maugeri et al., 2004).
Molecular Detection of Antimicrobial Resistance Gene Determinants
All of the isolates were subjected to molecular characterization of important β-lactamases in the Enterobacteriaceae family using multiplex, and duplex PCR protocols with the aid of a Biorad Thermal Cycler (United States), under conditions defined by Dallenne and colleagues (Dallenne et al., 2010). The various resistance genes encoding the different β-lactamases variants assayed were; blaTEM (comprising TEM-2 and TEM-1), blaCTX-M-9 (comprising CTX-M-14 and CTX-M-9), blaSHV (SHV-1), blaCTX-M-1 (comprising CTX-M-15, CTX-M-1, and CTX-M-3), blaCTX-M-2 (comprising CTX-M-2), blaPER (comprising PER-3 and PER-1), blaVEB (containing VEB-1 through VEB-6), blaGES (including GES-11 and GES-1 through GES-9), blaIMP (excluding IMP-25, IMP-22, IMP-18, IMP-16, and IMP-9), blaOXA-1-like (comprising OXA-30, OXA-4, and OXA-1), blaVIM (including VIM-2 and VIM-1), blaKPC (containing KPC-1 to KPC-5), and blaOXA-48-like. In the same vein, pAmpC-type β-lactamase variants mediated by plasmids, namely CIT, ACC, EBC, DHA, MOX, and FOX, were also assayed. They were organized into the following various categories. The blaCIT (BIL-1, CMY-12 to CMY-18, CMY-2 to CMY-7, LAT-1 to LAT-3, and CMY-21 to CMY-23), blaACC (including ACC-2 and ACC-1), blaEBC (MIR-1 and ACT-1), blaDHA (DHA-2 and DHA-1), blaMOX (CMY-19, MOX-1, CMY-1, MOX-2, and CMY-8 to CMY-11), and blaFOX (including FOX-1 to FOX-5). Genes encoding non-β-lactam resistance were investigated for all isolates as described previously (Titilawo et al., 2015) for tetracyclines (tetB, tetD, tetC, tetM, tetK, and tetA), sulphonamides (sulII and sulI), and phenicols (catII and catI). All primers were produced by Inqaba Biotechnological Industries (Pretoria, RSA). A volume of 5 µL of the amplified DNA was resolved in a 1.5% (w/v) agarose gel (Separations, RSA) stained with 5 µL ethidium bromide. A molecular marker of 100 base pairs (Thermo Scientific, United States) was used. The gel electrophoresis was run in 0.5X Tris-borate EDTA buffer for 1 h at 100 V and visualized in a UV transillumination (ALLIANCE 4.7, UVIec, Merton, London, United Kingdom).
Data Analysis
The data acquired were analyzed utilizing the descriptive statistical program available in Microsoft Excel 2010.
Results and Discussion
Total Quantification and Identification of Enterobacteriaceae in the Wastewater Treatment Plants
Water quality is a significant concern for many communities that rely on freshwater sources for consumption and recreation purposes. Concerns being raised regarding their pollution by pathogens (Sibanda et al., 2013) and on the requirement by law (Department of Water Affairs (DWAF), 2012), which states that treated effluents must be discharged into public streams or seas where the water was initially obtained. In this study, the average counts of the total Enterobacteriaceae bacteria were 3.37 × 104 CFU/100 ml and 5.75 × 105 CFU/100 ml in ADM and CHDM effluents, respectively. Since these effluents are being discharged into the surrounding surface water, the bacterial counts observed was compared with the compliance concentrations of the South African and WHO tolerance limits for effluents intended for discharge by municipal WWTPs into the receiving surface water. Properly treated effluent should not have a total bacterial count greater than 104 CFU/100 ml. The results obtained in this study is higher than the threshold of the bacterial count for municipal WWTP final effluents. These values are an indication that the effluents that are discharged into the surrounding surface water are of poor quality as enteric microorganisms are not adequately eliminated or reduced to acceptable limits. The results obtained are an indication that bacterial species are widely distributed in the WWTPs effluents. The quality of an effluent discharge is particularly an important criterium in assessing the efficiency of a WWTP. The surface water which receives these municipal WWTP effluents are being abstracted for irrigation, domestic, and religious activities (Fadare et al., 2020), thereby being a public health concern as they are unsuitable for domestic uses.
In total, 44 presumptive bacterial isolates were recovered from the different selective agar. These bacterial species belonged to four different families, which are Moraxellaceae (1), Morganellaceae (2), Staphylococcaceae (1), and Enterobacteriaceae (35). The remaining five bacterial species had a matching value lower than 1.7, which is the minimum score value for the reliability of the identity of an isolate using MALDI-TOF and were consequently excluded from this study. From the Enterobacteriaceae family, six genera were identified, which include Serratia (2), Proteus (4), Klebsiella (10), Escherichia (5), Enterobacter (8), and Citrobacter (6) as shown in Supplementary Table S1. The present study focuses on the last four genera of the Enterobacteriaceae family (n = 29). Among these, the most frequently detected bacterial species was K. pneumoniae (7/29; 24%), followed by E. cloacae (6/29; 21%) and E. coli (5/29; 17%). The other bacterial species identified were K. oxytoca and C. freundii (3/29; 10% each), C. koseri (2/29; 7%), and C. braakii, E. asburiae, and E. cancerogenus (1/29; 3% each), as shown in Figure 2. The detection and subsequent isolation of these Enterobacteriaceae indicate that the bacterial species were not eliminated during the various treatment stages in the WWTP. Enterobacteriaceae are members of the Proteobacteria phylum, which is distinguished by their capability of transferring DNA to bacteria from other classes. They can also promote genetic exchange between environmental and commensal bacteria in water and other environments (Ojer-Usoz et al., 2014). Different Enterobacteriaceae genera and species were found in this study, close to those found in the WWTPs of other nations, including Portugal, Colombia, and Spain (Ojer-Usoz et al., 2017; Aristizábal-Hoyos et al., 2019; Teixeira et al., 2020). These studies reported that members of the Enterobacteriaceae could survive the biological treatment processes employed in the management of municipal WWTPs.
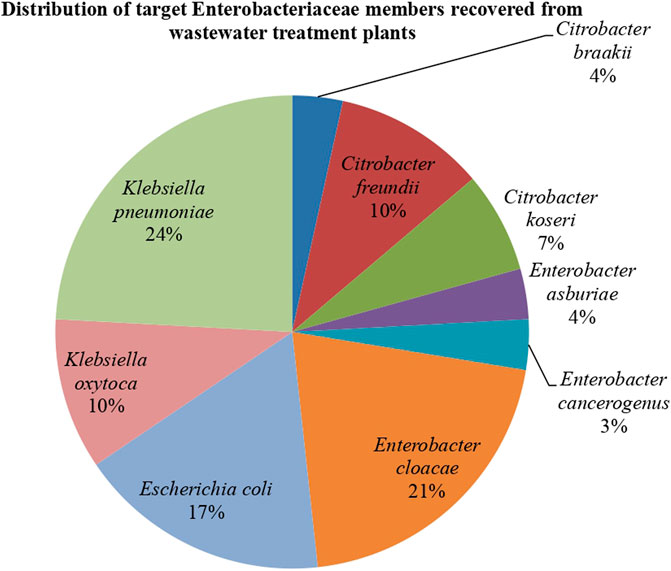
FIGURE 2. The distribution of the targeted Enterobacteriaceae’ members (n = 29) recovered from wastewater treatment plants.
Antibiogram Profiling of Targeted Enterobacteriaceae From Wastewater Treatment Plants
The antibiotic susceptibility profiles of the confirmed bacterial species recovered from final effluents of WWTPs are shown in Figure 3. Concerning the individual genera, all the Klebsiella spp. which were isolated exhibited resistance against three antibiotics. These are doxycycline, ampicillin, and cefotaxime which are from different antibiotic classes.
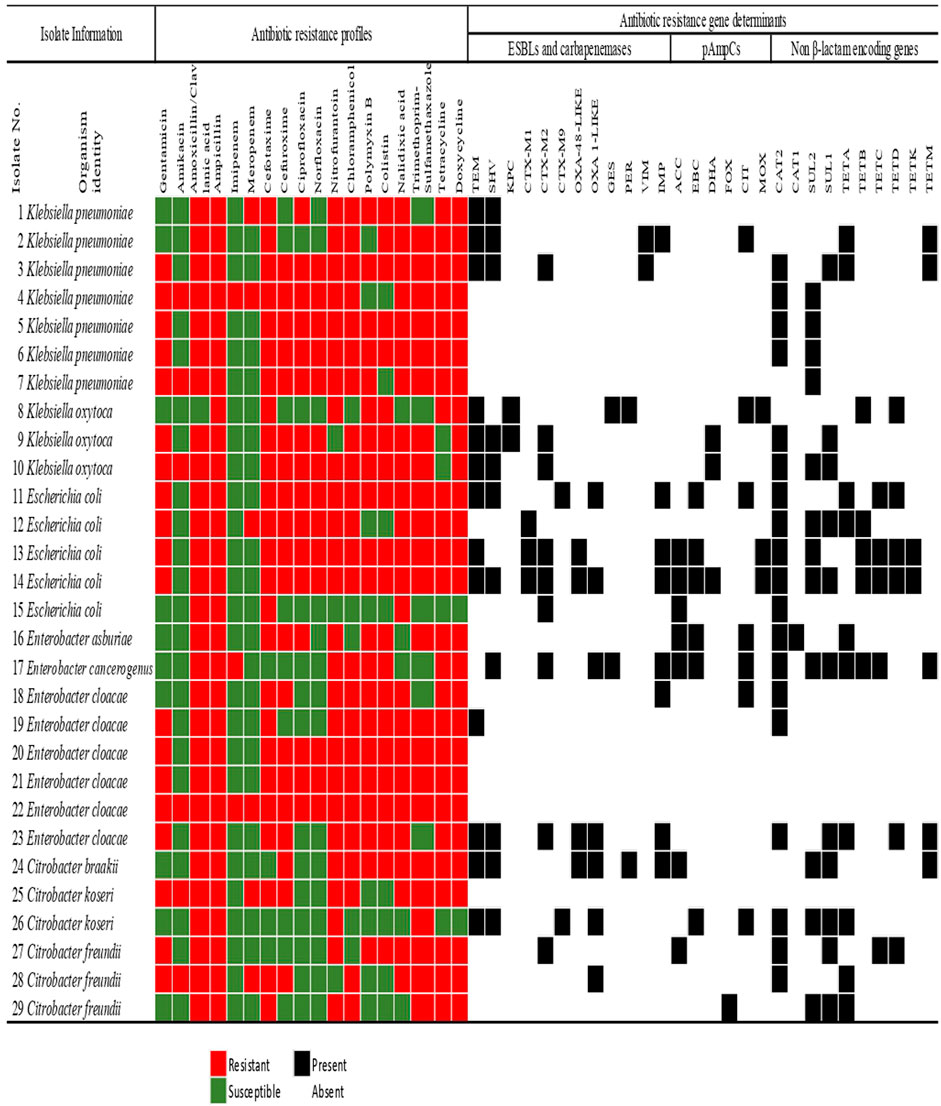
FIGURE 3. The Antimicrobial susceptibility profiling of members of the Enterobacteriaceae family recovered from municipal wastewater treatment plants indicating the extended-spectrum β-lactamase and carbapenemases, plasmid-mediated AmpC β-lactamase, and non-β-lactam resistance determinants. The colour code of the antimicrobial susceptibility profiles shows the phenotypes of the isolates to specific antibiotics in 11 different classes. The detection of the resistance gene determinants is indicated as present or absent.
In E. coli, all the isolates exhibited resistance against ampicillin, cefotaxime, amoxicillin/clavulanic acid, and nalidixic acid. However, none of the E. coli strains displayed resistance against imipenem or amikacin. All the species of Enterobacter were resistant to four different antibiotic classes. These classes were the β-lactams, polymyxins, tetracyclines, and nitrofurantoin. The least phenotypic resistance observed was against meropenem and amikacin, with only one E. cloacae strain (Isolate No. 26) exhibiting resistance against these antibiotics. Interestingly, 1 E. cloacae (Isolate No. 22) showed resistance against eighteen antibiotics assayed (Figure 3). All the Citrobacter spp. were resistant against amoxicillin-clavulanic acid, ampicillin, and trimethoprim-sulfamethoxazole but were all susceptible to imipenem, ciprofloxacin, and norfloxacin.
Generally, across all the Enterobacteriaceae members, some resistance was observed against all the antibiotics. The least resistance by the isolates was observed against amikacin and carbapenems. Carbapenems have been considered a drug of last resort. They are generally not prescribed as a front-line antimicrobial, so the low rate detection of resistance against these antimicrobials is unsurprising.
The Multiple Antibiotic Resistance Phenotype and Multiple Antibiotic Resistance Indice of Enterobacteriaceae From Wastewater Treatment Plants Effluents
The various MARP patterns and MARI displayed by the various genera of the Enterobacteriaceae are presented in Table 1. All the bacterial species displayed resistance against a minimum of three different classes of antibiotics out of the eleven classes assayed and were thus classified as MDR (Magiorakos et al., 2012). The average MARI obtained for all the isolates was 0.65, and it ranges from 0.22 to 1.0. Almost all the MARP patterns observed occurred singly except a pattern of resistance against 15 antibiotics observed in three E. coli strains. All the isolates have a MARI value greater than the benchmark of 0.2, indicating a higher than normal pressure of antibiotics in the WWTPs. Most of the antibiotics administered in the clinical or veterinary settings are incompletely broken down in the body and are thus excreted into the wastewater systems. Antibiotics in wastewater have been shown to exert selective pressure on antimicrobial-resistant bacterial organisms, promoting their dissemination into other habitats. (Li et al., 2019).
Generally, the results demonstrate that WWTPs are hotspots for shedding enteric pathogens with resistance to multiple classes of antibiotics into the receiving watersheds and, ultimately, into the environment. Many other researchers have reported this in their findings (Okoh et al., 2010; Korzeniewska and Harnisz, 2013; Adefisoye and Okoh, 2016; Igbinosa, 2016; Aristizábal-Hoyos et al., 2019). The results of the MAR index indicate that 41% (12/29) of the isolates recovered from the WWTPs exhibited resistance against a minimum of fifteen of the tested antibiotics. According to previous studies, wastewater, even after treatment, contains a proportion of different resistant bacterial populations (Huang et al., 2012; Ke et al., 2013; Amador et al., 2015; Guyomard-Rabenirina et al., 2017). The conditions in the WWTPs, according to these studies, are conducive to ARB proliferation, which could lead to the transmission of antibiotic-resistant genes to non-resistant bacteria.
Multiple Antibiotic Resistance Genotypes of Bacterial Species Recovered From Wastewater Treatment Plants’ Effluents
Findings from the antibiotic-resistant genes (ARGs) investigated in this study revealed that the isolates harboured various resistance genes responsible for the multiple phenotypic resistance displayed by the bacterial species. These resistance genes were broadly classified as the β-lactamases and the non-β-lactam resistance genes.
Out of all the Enterobacteriaceae investigated in the study, 86.2% (n = 25) harboured at least one of the various ARGs investigated. As indicated in Table 2, the MARG patterns of these microorganisms occurred uniquely. Three K. pneumoniae strains (Isolate No 4, 5, and 6) were positive to only two of the resistance genes assayed: catII and sulII only (Figure 3 and Table 2). Also, one K. pneumoniae strain (Isolate No. 7) harboured only one resistance gene (sulII) and was thus excluded from the MARG assessment. Twenty-two different genotypic patterns were observed in the MDR isolates carrying more than one resistance gene. The different MARGs show that each isolate has its own set of genes that could have been responsible for the various resistant phenotypes.
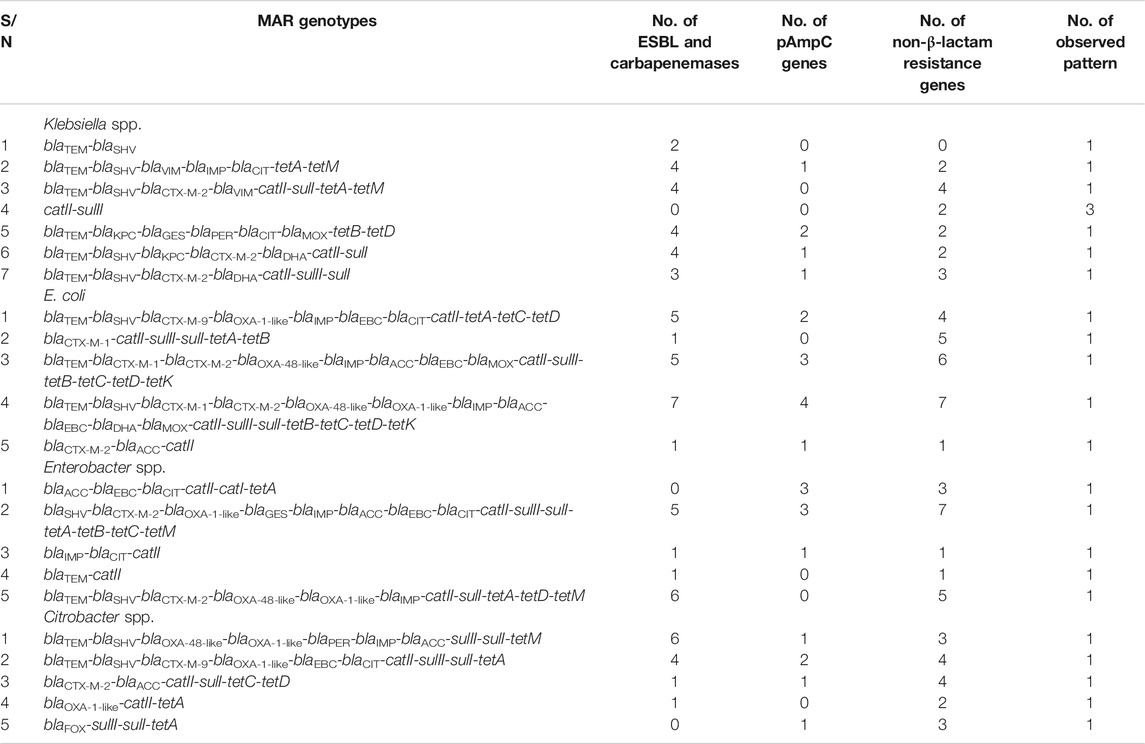
TABLE 2. The patterns of the various multiple antibiotic resistance genotypes of Enterobacteriaceae recovered from wastewater treatment plants effluents.
Among the four different genera considered, the E. coli harboured the most frequently detected resistance genes. Their higher rate of resistance than the other genera studied is attributable to their proclivity for constant possession and inclusion of ARGs, especially within the aquatic context. The ease with which ARGs can be acquired in E. coli has been documented in previous studies (Ahlstrom et al., 2018; Li et al., 2019).
As seen in Figure 3, all the isolates with multiple antimicrobial resistance genetic determinants exhibited a co-occurrence of the β-lactamases with the non-β-lactam encoding genes, except only one K. pneumoniae (Isolate No. 1) which harboured only two β-lactamases. The highest frequency of resistance genes detected was in an E. coli isolate (Isolate No. 14). It was followed by a total of fifteen various resistance genes detected in the only E. cancerogenus reported in this study. The detection of high ARGs in these enteric microorganisms in effluents discharged to surface water is a cause of concern. This is because some communities rely on surface water for their daily domestic activities such as bathing, and washing. Therefore, the introduction of these biological contaminants may pose a risk to public health and safety.
Various research has emphasized that contaminated water supplies are primarily liable for enhancing the dissemination of ARGs (Zhang et al., 2009; Wright, 2010; Tacão et al., 2012). The Enterobacteriaceae family, which includes common nosocomial and opportunistic pathogens as well as natural commensals of warm-blooded animals, is known to play a key function in the spread of ARGs from environmental microorganisms to other species, ultimately reaching human pathogens (Machado et al., 2009; Tacão et al., 2012).
β-lactamases Detected in Effluents of Wastewater Treatment Plants
Generally, it was found that 72.4% (n = 21) of the Enterobacteriaceae analyzed harboured a minimum of one of the β-lactamase resistance genes assessed. From the 92 β-lactamases detected, 70.7% (n = 65) corresponds to the ESBLs and carbapenemases while the remaining 29.3% (n = 27) belonged to the pAmpCs. Klebsiella spp. harboured most of the former categories with 32.3% (n = 21) detected, and this was keenly followed by E. coli at 29.2% (n = 19). The Enterobacter spp. and Citrobacter spp. also harboured 20% (n = 13) and 18.5% (n = 12).
For the ESBLs, the CTX-M group was found the most frequently at 21.5% (n = 14) with distribution as follows; blaCTX-M-2 group 13.8% (n = 9), blaCTX-M-1 group 4.6% (n = 3), and blaCTX-M-9 group 3.1% (n = 2). The next dominant category of the ESBLs detected was blaTEM with 20% (n = 13), followed by blaSHV with 16.9% (n = 11). Other β-lactamase variants detected were blaIMP with 12.3% (n = 8), blaOXA-1-like with 10.8% (n = 7), blaOXA-48-like with 6.1% (n = 4). The variants with the lowest frequency were detected at 3.1% (n = 2), and these include blaPER,blaVIMblaKPC, and blaGES (Figure 4A). Regarding the distribution of the β-lactamases by microorganisms, the predominance of blaTEM, blaSHV, blaCTX-M-2 group, and blaIMP can be noted in all bacterial genera. A detailed breakdown of the percentages of the various genotypes detected with the corresponding number of each gene detected across the bacterial genus is presented in Figure 4A.
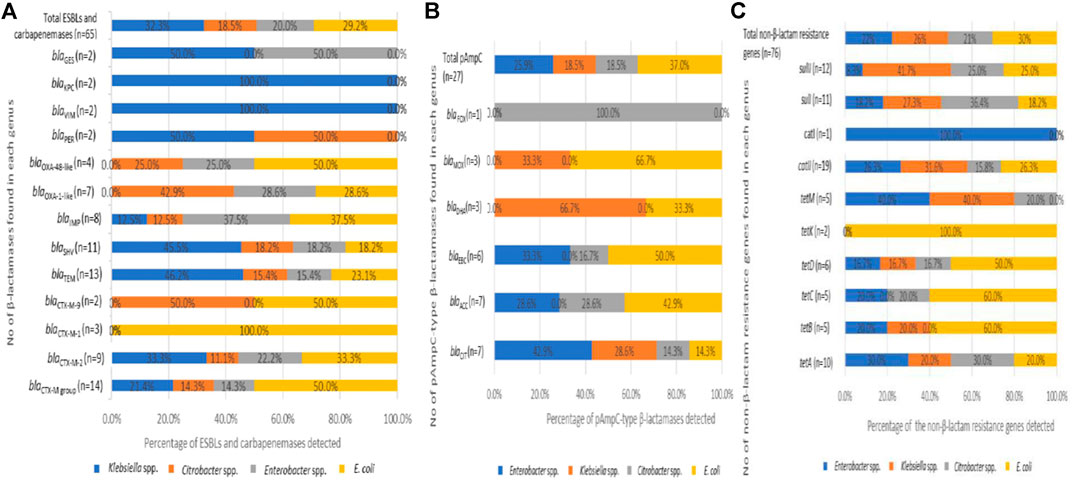
FIGURE 4. The distribution of extended-spectrum β-lactamases, plasmid-mediated AmpC β-lactamases in the Enterobacteriaceae recovered from the final effluents of WWTPs. (A) Distribution of ESBLs (B) Distribution of pAmpCs and (C) Distribution of non-β-lactam resistance genes which are present in the recovered bacterial species.
Initially, the most common β-lactamase enzyme is the TEM-1 that could hydrolyze penicillins among the Enterobacteriaceae. Soon after, the blaTEM spread rapidly to other species and, with a few changes in their amino acid components, gave rise to blaSHV enzymes, which hydrolyze cephalosporins. However, the blaSHV were more frequently detected in Klebsiella spp. than the blaTEM (Datta and Kontomichalou, 1965; Brunton et al., 1986; Bush and Jacoby, 2010; Aristizábal-Hoyos et al., 2019). As per this study, the highest frequency of blaTEM, 46.2% (n = 6) and blaSHV, 45.5% (n = 5), were recorded among the Klebsiella spp. (Figure 4A). The Klebsiella spp. harbouring β-lactamases such as the blaTEM and blaSHV have been previously reported to cause nosocomial infections (Rawat and Nair, 2010).
Since the detection of another β-lactamase enzyme, blaCTX-M-1, recovered in an E. coli isolate (Bauernfeind et al., 1990; Bush and Jacoby, 2010), it has been known to spread rapidly in different bacterial species. The ability of the blaCTX-M group to spread rapidly in other Enterobacteriaceae has been established (Poirel et al., 2002; Aristizábal-Hoyos et al., 2019), so this makes the rate of detection of the blaCTX-M higher than the earlier detected enzymes blaTEM and blaSHV. The blaCTX-M group has been reported as the most predominant enzymes among the β-lactamases, followed by the blaTEM (Tham, 2010; Ahlstrom et al., 2018; Aristizábal-Hoyos et al., 2019). This report is consistent with the predominant detection of the blaCTX-M group (21.5%) keenly followed by the blaTEM (20%) detected in this study. Some other researchers have also described the dominance of the blaCTX-M variants in many different surface water (Zarfel et al., 2013; Zurfluh et al., 2013; Ojer-Usoz et al., 2014). Also, the blaCTX-M variants are among the most widespread in South Africa (Njage and Buys, 2015; Fadare et al., 2020). Interestingly, we discovered isolates from various genera such as E. cancerogenus, E. cloacae, C. koseri, C. freundii, K. oxytoca, K. pneumoniae, and E. coli harbouring the various blaCTX-M variants. It emphasizes the significance of the research into the existence of cephalosporinases in WWTP final effluents. This study's finding of the highest frequency of blaCTX-M genes in E. coli confirms previous findings. Also, the presence of ESBL in this genus is common, as these enzymes impart resistance to cephalosporins, one of the most commonly used antibiotic classes in clinical practice (Shi et al., 2015; Borgogna et al., 2016; Aristizábal-Hoyos et al., 2019). The blaTEM gene is also one of the most frequently identified genes conferring resistance to β-lactams. It has been shown to be generally discovered in effluents of WWTPs owing to its ability to evade the wastewater treatment stages and, consequently, be detected in rivers downstream of WWTPs (Yu et al., 2018; Aristizábal-Hoyos et al., 2019). The finding of the various ESBLs variants, especially the dominance of blaTEM, blaSHV, and blaCTX-M, in all the bacterial genera recovered from the treated final effluent of these WWTPs, is important. It appears that the traditional biological treatment used in these WWTPs selectively favours the development of antimicrobial resistance, which may have arisen on account of interactions amid susceptible and resistant microorganisms, as well as the continual selection pressure exerted by antibiotics at subinhibitory concentrations (Aristizábal-Hoyos et al., 2019).
In the other category of the β-lactamases assayed, the pAmpCs resistant gene determinants were also detected in this study. The highest frequency of the total pAmpC variants detected was harboured in E. coli with 37% (n = 10). In this category, the most predominant was blaCIT and blaACC with 25.9% (n = 7) each. The blaFOX was detected in only one C. freundii (Isolate No. 24), as seen in Figure 3. The percentage frequencies of these pAmpC-type resistance genes are as indicated in Figure 4B. Globally, the frequencies of the detection of the pAmpCs have been reported to be lower than their ESBL counterpart (Aristizábal-Hoyos et al., 2019). The detection of plasmid-mediated resistance genes in this study shows that they are not only restricted to the clinical settings but are also detected even in the environment.
In comparison to other studies, the prevalence of the pAmpCs reported in this study varied. In the final effluent of WWTPs in Colombia, only two categories of pAmpCs were detected in E. cloacae, E. coli, and C. freundii. Here, the incidence of LAT/BIL/CMY belonging to the CIT group was similar (n = 7) to that observed in our study, while the incidence of ACT/MIR belonging to the EBC group (n = 1) was relatively lower. Suffice to say that comparing antibiotic resistances in microorganisms across studies is a good predictor of incidence. However, it does not prove real disparities in incidence due to differences in sample forms, strain isolation methods, or resistance testing approaches (Njage and Buys, 2015). However, even a low prevalence of pAmpCs in Enterobacteriaceae is a cause for concern because clinical specimens have been shown to have elevated levels of AmpC β-lactamase expression. A significant threat is the increased development of chromosomal AmpC β-lactamases associated with the possession of plasmid-mediated AmpC-lactamases (Jacoby, 2009).
In the Enterobacteriaceae group, the principal means of antimicrobial resistance is the ability to produce β-lactamases. These enzymes break the chemical structural backbone, the β-lactam ring, of the largest and commonly used antibiotic classes (Bush and Jacoby, 2010). From the results of this study indicated in Table 2, these isolates' ability to possess these β-lactamases is quite concerning. This scenario implies that existing antibiotics’ efficacy could be undermined while also hindering the development of new ones.
Non-β-Lactam Resistance Encoding Genes Detected in Effluents of Wastewater Treatment Plants
Plasmids carrying genes responsible for various β-lactamases usually encode resistance against other antimicrobial classes due to their large sizes (Jacoby, 2009). In this study, it was found that 86.2% (n = 25) of the Enterobacteriaceae analyzed have a minimum of one of the non-β-lactam resistance encoding genes examined. The various ARGs against the antibiotic class tetracyclines was the most frequently detected at 43.4% (n = 33). The resistance against the sulphonamide class was 30.3% (n = 23), while the resistance against the antibiotic class phenicols was 26.3% (n = 20). The frequency of each of the non-β-lactam resistance determinants across the Enterobacteriaceae investigated includes tetA (13.2%), tetD (7.9%), tetB, tetC, and tetM (6.6% each), catII (25%), catI (1.3%), sulII (15.8%), and sulI (14.5%).
E. coli harboured the highest resistance genes detected at the genus level with 30.3% (n = 23). It was represented in almost all the categories of the genes assayed except tetM and catI. E. coli harboured the only tetK detected. The percentage frequency of detecting each of the non-β-lactam resistance genes across the various bacterial genus is shown in Figure 4. The high rate of detection of non-β-lactam resistance genes observed is attributable to the fact that these antibiotic classes are commonly administered as first-line antimicrobials in clinical and veterinary settings.
Conclusion
The study revealed the presence of MDR Enterobacteriaceae’s members in the final effluents of WWTPs along with a repertoire of various important clinically relevant β-lactamases, and other essential non-β-lactam resistance genes that can be easily transferred to other species in a variety of niches. This diversity confirms the selection pressure present in the WWTPs, implying that this type of environment favours the development and widespread of ARB and ARGs due to the dissemination of plasmid-encoded enzymes capable of hydrolyzing essential antibiotics. It also shows that, even when functioning at optimal capacity, the current design of the studied WWTPs do not enable them to prevent the spread of ARB and ARGs into the environment, posing a serious public health danger. In developing countries like South Africa, where surface water is used for domestic and irrigation purposes the poor quality of effluent discharged from WWTPs is a major concern for public health safety. Hence, this study reveals that concerted efforts should be made to improve WWTP's processes to include monitoring the presence of ARGs in wastewater before they are released into the environment. The blaCTX-M genes which was widely detected in this study and other similar studies may be used as a marker to monitor the spread of resistance and mitigate the risk on human health.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
Conceptualization, AO and FF; methodology, AO and FF; investigation, FF; data curation, FF; writing—original draft preparation, FF; writing—review and editing, AO and FF; supervision, AO; funding acquisition, AO. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the South African Medical Research Council’s financial support, the National Research Foundation (NRF) and the Partnerships for Enhanced Engagement in Research (PEER), a USAID/DST programme. These funding agencies did not partake in the study’s design, and conclusions arrived herein are entirely those of the authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.711950/full#supplementary-material
References
Adefisoye, M. A., and Okoh, A. I. (2016). Identification and Antimicrobial Resistance Prevalence of Pathogenic Escherichia Coli Strains from Treated Wastewater Effluents in Eastern Cape, South Africa. MicrobiologyOpen 5 (1), 143–151. doi:10.1002/mbo3.319
Ahlstrom, C. A., Bonnedahl, J., Woksepp, H., Hernandez, J., Olsen, B., Ramey, A. M., et al. (2018). Acquisition and Dissemination of Cephalosporin-Resistant E. Coli in Migratory Birds Sampled at an Alaska Landfill as Inferred through Genomic Analysis. Sci. Rep. 8 (1), 1–11. doi:10.1038/s41598-018-25474-w
Amador, P. P., Fernandes, R., Prudêncio, M. C., Barreto, M. P., Duarte, I. M., and Isabel, M. (2015). Antibiotic Resistance in Wastewater: Occurrence and Fate ofEnterobacteriaceaeproducers of Class A and Class C β-lactamases. J. Environ. Sci. Health A 50 (1), 26–39. doi:10.1080/10934529.2015.964602
Aristizábal-Hoyos, A. M., Rodríguez, E. A., Arias, L., and Jiménez, J. N. (2019). High Clonal Diversity of Multidrug-Resistant and Extended Spectrum Beta-Lactamase-Producing Escherichia Coli in a Wastewater Treatment Plant. J. Environ. Manage. 245 (September), 37–47. doi:10.1016/j.jenvman.2019.05.073
Ateba, C. N., and Bezuidenhout, C. C. (2008). Characterisation of Escherichia Coli O157 Strains from Humans, Cattle and Pigs in the North-West Province, South Africa. Int. J. Food Microbiol. 128 (2), 181–188. doi:10.1016/j.ijfoodmicro.2008.08.011
Bauernfeind, A., Schweighart, S., and Grimm, H. (1990). A New Plasmidic Cefotaximase in a Clinical Isolate of Escherichia Coli. Infection 18 (5), 294–298. doi:10.1007/BF01647010
Borgogna, T. R., Borgogna, J.-L., Mielke, J. A., Brown, C. J., Top, E. M., Botts, R. T., et al. (2016). High Diversity of CTX-M Extended-Spectrum β-Lactamases in Municipal Wastewater and Urban Wetlands. Microb. Drug Resist. 22 (4), 312–320. doi:10.1089/mdr.2015.0197
Brunton, J., Clare, D., and Meier, M. A. (1986). Molecular Epidemiology of Antibiotic Resistance Plasmids of Haemophilus Species and Neisseria Gonorrhoeae. Clin. Infect. Dis. 8 (5), 713–724. doi:10.1093/clinids/8.5.713
Bush, K., and Jacoby, G. A. (2010). Updated Functional Classification of β-Lactamases. Antimicrob. Agents Chemother. 54 (3), 969–976. doi:10.1128/AAC.01009-09
Cantón, R., Akóva, M., Carmeli, Y., Giske, C. G., Glupczynski, Y., Gniadkowski, M., et al. (2012). Rapid Evolution and Spread of Carbapenemases Among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18 (5), 413–431. doi:10.1111/j.1469-0691.2012.03821.x
CLSI (2018). M100-S23 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Information Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
Dallenne, C., da Costa, A., Decré, D., Favier, C., and Arlet, G. (2010). Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65 (3), 490–495. doi:10.1093/jac/dkp498
Datta, N., and Kontomichalou, P. (1965). Penicillinase Synthesis Controlled by Infectious R Factors in Enterobacteriaceae. Nature 208 (5007), 239–241. doi:10.1038/208239a0
Department of Water Affairs (DWAF) (2012). 2012 Green Drop Progress Report. Pretoria, Republic of South Africa: Department of Water AffairsAvailable at: http://www.dwa.gov.za/documents/AnnualReports/DWA AnnualReport 2012-13.pdf.
Fadare, F. T., Adefisoye, M. A., and Okoh, A. I. (2020). Occurrence, Identification and Antibiogram Signatures of Selected Enterobacteriaceae from Tsomo and Tyhume Rivers in the Eastern Cape Province, Republic of South Africa. PLoS ONE 08 (11), 246025. doi:10.1101/2020.08.11.246025
Guyomard-Rabenirina, S., Dartron, C., Falord, M., Sadikalay, S., Ducat, C., Richard, V., et al. (2017). Resistance to Antimicrobial Drugs in Different Surface Waters and Wastewaters of Guadeloupe. PLoS ONE 12 (3), e0173155–17. doi:10.1371/journal.pone.0173155
Huang, J.-J., Hu, H.-Y., Lu, S.-Q., Li, Y., Tang, F., Lu, Y., et al. (2012). Monitoring and Evaluation of Antibiotic-Resistant Bacteria at a Municipal Wastewater Treatment Plant in China. Environ. Int. 42 (July), 31–36. doi:10.1016/j.envint.2011.03.001
Igbinosa, E. O. (2016). Detection and Antimicrobial Resistance ofVibrioIsolates in Aquaculture Environments: Implications for Public Health. Microb. Drug Resist. 22 (3), 238–245. doi:10.1089/mdr.2015.0169
Jacoby, G. A. (2009). AmpC β-Lactamases. Clin. Microbiol. Rev. 22 (1), 161–182. doi:10.1128/CMR.00036-08
Korzeniewska, E., and Harnisz, M. (2013). Beta-Lactamase-Producing Enterobacteriaceae in Hospital Effluents. J. Environ. Manage. 123 (July), 1–7. doi:10.1016/j.jenvman.2013.03.024
Kotlarska, E., Łuczkiewicz, A., Pisowacka, M., and Burzyński, A. (2015). Antibiotic Resistance and Prevalence of Class 1 and 2 Integrons in Escherichia Coli Isolated from Two Wastewater Treatment Plants, and Their Receiving Waters (Gulf of Gdansk, Baltic Sea, Poland). Environ. Sci. Pollut. Res. 22 (3), 2018–2030. doi:10.1007/s11356-014-3474-7
Krumperman, P. H. (1983). Multiple Antibiotic Resistance Indexing of Escherichia coli to Identify High-Risk Sources of Fecal Contamination of Foods. Appl. Environ. Microbiol. 46, 165–170. doi:10.1128/aem.46.1.165-170.1983
Lamprecht, C., Romanis, M., Huisamen, N., Carinus, A., Schoeman, N., Sigge, G. O., et al. (2014). Escherichia Coli with Virulence Factors and Multidrug Resistance in the Plankenburg River. Sajs 110 (9–10), 1–6. doi:10.1590/sajs.2014/20130347
Li, Q., Chang, W., Zhang, H., Hu, D., and Wang, X. (2019). The Role of Plasmids in the Multiple Antibiotic Resistance Transfer in ESBLs-Producing Escherichia Coli Isolated from Wastewater Treatment Plants. Front. Microbiol. 10 (APR), 1–8. doi:10.3389/fmicb.2019.00633
Machado, E., Coque, T. M., Cantón, R., Sousa, J. C., Silva, D., Ramos, M., et al. (2009). Leakage into Portuguese Aquatic Environments of Extended-Spectrum- -Lactamase-Producing Enterobacteriaceae. J. Antimicrob. Chemother. 63 (3), 616–618. doi:10.1093/jac/dkn510
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 18 (3), 268–281. doi:10.1111/j.1469-0691.2011.03570.x
Maugeri, T. L., Carbone, M., Fera, M. T., Irrera, G. P., and Gugliandolo, C. (2004). Distribution of Potentially Pathogenic Bacteria as Free Living and Plankton Associated in a Marine Coastal Zone. J. Appl. Microbiol. 97 (2), 354–361. doi:10.1111/j.1365-2672.2004.02303.x
Njage, P. M. K., and Buys, E. M. (2015). Pathogenic and Commensal E Scherichia Coli from Irrigation Water Show Potential in Transmission of Extended Spectrum and AmpC β‐lactamases Determinants to Isolates from Lettuce. Microb. Biotechnol. 8 (3), 462–473. doi:10.1111/1751-7915.12234
Ojer-Usoz, E., González, D., and Vitas, A. I. (2017). Clonal Diversity of ESBL-Producing Escherichia Coli Isolated from Environmental, Human and Food Samples. Int. J. Environ. Res. Public Health 14 (7), 676. doi:10.3390/ijerph14070676
Ojer-Usoz, E., González, D., García-Jalón, I., and Vitas, A. I. (2014). High Dissemination of Extended-Spectrum β-lactamase-producing Enterobacteriaceae in Effluents from Wastewater Treatment Plants. Water Res. 56, 37–47. doi:10.1016/j.watres.2014.02.041
Okoh, A. I., Igbinosa, E. O., and O Igbinosa, Etinosa. (2010). Antibiotic Susceptibility Profiles of Some Vibrio Strains Isolated from Wastewater Final Effluents in a Rural Community of the Eastern Cape Province of South Africa. BMC Microbiol. 10 (May), 143. doi:10.1186/1471-2180-10-143
Osundiya, O., Oladele, R., and Oduyebo, O. (2013). Multiple Antibiotic Resistance (MAR) Indices of Pseudomonas and Klebsiella Species Isolates in Lagos University Teaching Hospital. Af J. Clin. Exp. Micro 14 (3), 164–168. doi:10.4314/ajcem.v14i3.8
Paterson, D. L., and Bonomo, R. A. (2005). Extended-Spectrum β-Lactamases: a Clinical Update. Clin. Microbiol. Rev. 18 (4), 657–686. doi:10.1128/CMR.18.4.657-686.2005
Paterson, D. L. (2000). Recommendation for Treatment of Severe Infections Caused by Enterobacteriaceae Producing Extended-Spectrum β-lactamases (ESBLs). Clin. Microbiol. Infect. 6 (9), 460–463. doi:10.1046/j.1469-0691.2000.00107.x
Poirel, L., Kämpfer, P., and Nordmann, P. (2002). Chromosome-Encoded Ambler Class A β-Lactamase of Kluyvera Georgiana , a Probable Progenitor of a Subgroup of CTX-M Extended-Spectrum β-Lactamases. Antimicrob. Agents Chemother. 46 (12), 4038–4040. doi:10.1128/aac.46.12.4038-4040.2002
Rawat, D., and Nair, D. (2010). Extended-spectrum SS-Lactamases in Gram Negative Bacteria. J. Glob. Infect Dis 2 (3), 263. doi:10.4103/0974-777x.68531
Shi, H., Sun, F., Chen, J., Ou, Q., Feng, W., Yong, X., et al. (2015). Epidemiology of CTX-M-type Extended-Spectrum Beta-Lactamase (ESBL)-Producing Nosocomial -Escherichia Coli Infection in China. Ann. Clin. Microbiol. Antimicrobials 14 (January), 4. doi:10.1186/s12941-015-0063-7
Sibanda, T., Chigor, V. N., and Okoh, A. I. (2013). Seasonal and Spatio-Temporal Distribution of Faecal-Indicator Bacteria in Tyume River in the Eastern Cape Province, South Africa. Environ. Monit. Assess. 185 (8), 6579–6590. doi:10.1007/s10661-012-3048-4
Standard Methods (2005). Standard Methods for the Examination of Water and Wastewater. 20th Edn. American Public Health Association (APHA)): Washington DC, USA. System and Intestinal Colonisation. Proc. Natl. Acad. Sci. 90, 3750–3754.
Standing, T.-A., Du Plessis, E., Duvenage, S., and Korsten, L. (2013). Internalisation Potential of Escherichia Coli O157:H7, Listeria Monocytogenes, Salmonella Enterica Subsp. Enterica Serovar Typhimurium and Staphylococcus Aureus in Lettuce Seedlings and Mature Plants. J. Water Health 11 (2), 210–223. doi:10.2166/wh.2013.164
Tärnberg, Maria. (2012). Extended-Spectrum Beta-Lactamase Producing Enterobacteriaceae : Aspects on Detection, Epidemiology and Multi-Drug ResistanceLinköping University.
Tacão, M., Correia, A., and Henriques, I. (2012). Resistance to Broad-Spectrum Antibiotics in Aquatic Systems: Anthropogenic Activities Modulate the Dissemination ofblaCTX-M-like Genes. Appl. Environ. Microbiol. 78 (12), 4134–4140. doi:10.1128/AEM.00359-12
Teixeira, P., Tacão, M., Pureza, L., Gonçalves, J., Silva, A., Cruz-Schneider, M. P., et al. (2020). Occurrence of Carbapenemase-Producing Enterobacteriaceae in a Portuguese River: BlaNDM, BlaKPC and BlaGES Among the Detected Genes. Environ. Pollut. 260, 113913. doi:10.1016/j.envpol.2020.113913
Tham, Johan. (2010). Extended-Spectrum Beta-Lactamase Producing-Enterobacteriaceae: Epidemiology, Risk Factors, and Duration of Carriage. Arch. Pediatr. 17 (Suppl. 4).
Titilawo, Y., Obi, L., and Okoh, A. (2015). Antimicrobial Resistance Determinants of Escherichia Coli Isolates Recovered from Some Rivers in Osun State, South-Western Nigeria: Implications for Public Health. Sci. Total Environ. 523, 82–94. doi:10.1016/j.scitotenv.2015.03.095
WHO (2014). Antimicrobial Resistance: Global Report on Surveillance 2014. Available at: https://www.who.int/drugresistance/documents/surveillancereport/en/ (Accessed April 28, 2021).
WHO (2017). Critically Important Antimicrobials for Human Medicine. Geneva: World Health Organization.
Wright, G. D. (2010). Antibiotic Resistance in the Environment: A Link to the Clinic? Curr. Opin. Microbiol. 13 (5), 589–594. doi:10.1016/j.mib.2010.08.005
Yu, X., Zhang, M., Zuo, J., Shi, X., Tang, X., Chen, L., et al. (2018). Evaluation of Antibiotic Resistant Lactose Fermentative Opportunistic Pathogenic Enterobacteriaceae Bacteria and blaTEM-2 Gene in Cephalosporin Wastewater and its Discharge Receiving River. J. Environ. Manage. 228 (December), 458–465. doi:10.1016/j.jenvman.2018.09.043
Zarfel, G., Galler, H., Feierl, G., Haas, D., Kittinger, C., Leitner, E., et al. (2013). Comparison of Extended-Spectrum-β-Lactamase (ESBL) Carrying Escherichia coli from Sewage Sludge and Human Urinary Tract Infection. Environ. Pollut. 173, 192–199. doi:10.1016/j.envpol.2012.09.019
Zhang, X.-X., Zhang, T., Fang, H. H. P., and Fang, P. (2009). Antibiotic Resistance Genes in Water Environment. Appl. Microbiol. Biotechnol. 82 (3), 397–414. doi:10.1007/s00253-008-1829-z
Keywords: wastewater treatment plant, extended—spectrum β—lactamases, plasmid-mediated AmpC beta-lactamase, enterobacteriaceae, multidrug-resistance
Citation: Fadare FT and Okoh AI (2021) The Abundance of Genes Encoding ESBL, pAmpC and Non-β-Lactam Resistance in Multidrug-Resistant Enterobacteriaceae Recovered From Wastewater Effluents. Front. Environ. Sci. 9:711950. doi: 10.3389/fenvs.2021.711950
Received: 19 May 2021; Accepted: 12 July 2021;
Published: 23 July 2021.
Edited by:
Ilunga Kamika, University of South Africa, South AfricaReviewed by:
Mohamed Salah Abbassi, Tunis El Manar University, TunisiaNaga Raju Maddela, Technical University of Manabi, Ecuador
Copyright © 2021 Fadare and Okoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Folake Temitope Fadare, folakefadare2002@gmail.com
 Folake Temitope Fadare
Folake Temitope Fadare Anthony Ifeanyi Okoh1,2
Anthony Ifeanyi Okoh1,2