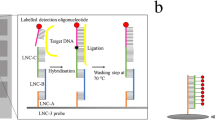
Abstract
In pathogen diagnostics, conventional culture-based assays remain the gold-standard; however, they are time-consuming, and shows the low positivity rate. Prior treatment with antibiotics is one of the major factors lowering the culture positivity rate. It is therefore important to evaluate the effects of prior antibiotic treatment on established detection methods and to develop sensitive and specific methods for detecting pathogens. Here, we report the detection of bacteria in samples containing antibiotics. Escherichia coli and Klebsiella pneumoniae were chosen as model and they were independently inoculated into culture flasks along with ceftriaxone (0.5× MIC, 1× MIC, 2× MIC). After 0, 2, 4, 6, 12, and 24 h, samples were collected from each flask and divided into two for evaluation using microarray or BacT/Alert culture-based systems. The newly designed probes showed a detection limit of 101 CFU for E. coli and 102 CFU for K. pneumoniae with high specificities. DNA microarray obtained true positive results at approximately 4 to 24 h after antimicrobial treatment, in which the culture-based method failed to detect the pathogenic bacteria. The DNA microarray-based assay could be useful for efficient detection of pathogenic bacteria in a clinical setting, allowing for appropriate administration of antibiotics in infected patients.
Similar content being viewed by others
References
Reimer, L. G., M. L. Wilson, and M. P. Weinstein (1997) Update on detection of bacteremia and fungemia. Clin. Microbiol. Rev. 10: 444–465.
Yoo, S. M. and S. Y. Lee (2010) DNA microarray for the identification of pathogens causing bloodstream infections. Expert. Rev. Mol. Diagn. 10: 263–268.
Bouza, E., D. Sousa, M. Rodríguez-Créixems, J. G. Lechuz, and P. Muñoz (2007) Is the volume of blood cultured still a significant factor in the diagnosis of bloodstream infections? J. Clin. Microbiol. 45: 2765–2769.
Lamy, B., S. Dargere, M. C. Arendrup, J. J. Parienti, and P. Tattevin (2016) How to optimize the use of blood cultures for the diagnosis of bloodstream infections? A state-of-the art. Front. Microbiol. 7: 697.
Al-Mayahi, M., A. Cian, B. A. Lipsky, D. Suvà, C. Müller, C. Landelle, H. H. Miozzari, and I. Uçkay (2015) Administration of antibiotic agents before intraoperative sampling in orthopedic infections alters culture results. J. Infect. 71: 518–525.
Bidell, M. R., M. P. Opraseuth, M. Yoon, J. Mohr, and T. P. Lodise (2017) Effect of prior receipt of antibiotics on the pathogen distribution and antibiotic resistance profile of key Gram-negative pathogens among patients with hospital-onset urinary tract infections. BMC Infect. Dis. 17: 176.
Kim, C. J., K. H. Song, W. B. Park, E. S. Kim, S. W. Park, H. B. Kim, M. D. Oh, and N. J. Kim (2012) Microbiologically and clinically diagnosed vertebral osteomyelitis: impact of prior antibiotic exposure. Antimicrob. Agents Chemother. 56: 2122–2124.
Shahi, A., C. Deirmengian, C. Higuera, A. Chen, C. Restrepo, B. Zmistowski, and J. Parvizi (2015) Premature therapeutic antimicrobial treatments can compromise the diagnosis of late periprosthetic joint infection. Clin. Orthop. Relat. Res. 473: 2244–2249.
Bearman, G. M. L. and R. P. Wenzel (2005) Bacteremias: a leading cause of death. Arch. Med. Res. 36: 646–659.
Peach, B. C. (2017) Implications of the new sepsis definition on research and practice. J. Crit. Care. 38: 259–262.
Singer, M., C. S. Deutschman, C. W. Seymour, M. Shankar-Hari, D. Annane, M. Bauer, R. Bellomo, G. R. Bernard, J. D. Chiche, C. M. Coopersmith, R. S. Hotchkiss, M. M. Levy, J. C. Marshall, G. S. Martin, S. M. Opal, G. D. Rubenfeld, T. van der Poll, J. L. Vincent, and D. C. Angus (2016) The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 315: 801–810.
Timsit, J. F., J. F. Soubirou, G. Voiriot, S. Chemam, M. Neuville, B. Mourvillier, R. Sonneville, E. Mariotte, L. Bouadma, and M. Wolff (2014) Treatment of bloodstream infections in ICUs. BMC Infect. Dis. 14: 489.
Flayhart, D., A. P. Borek, T. Wakefield, J. Dick, and K. C. Carroll (2007) Comparison of BACTEC PLUS blood culture media to BacT/Alert FA blood culture media for detection of bacterial pathogens in samples containing therapeutic levels of antibiotics. J. Clin. Microbiol. 45: 816–821.
Kang, H. and S. Kim (2012) Clinical features associated with blood cultures according to the use of antimicrobial agents prior to blood collection. Ann. Clin. Microbiol. 15: 21–26.
Templier, V., T. Livache, S. Boisset, M. Maurin, S. Slimani, R. Mathey, and Y. Roupioz (2017) Biochips for direct detection and identification of bacteria in blood culture-like conditions. Sci. Rep. 7: 9457.
Lovern, D., B. Katzin, K. Johnson, D. Broadwell, E. Miller, A. Gates, P. Deol, K. Doing, A. van Belkum, C. Marshall, E. Mathias, and W. M. Dunne Jr. (2016) Antimicrobial binding and growth kinetics in BacT/ALERT® FA Plus and BACTEC® Aerobic/F Plus blood culture media. Eur. J. Clin. Microbiol. Infect. Dis. 35: 2033–2036.
Doern, C. D., S. Mirrett, D. Halstead, J. Abid, P. Okada, and L. B. Reller (2014) Controlled clinical comparison of new pediatric medium with adsorbent polymeric beads (PF Plus) versus charcoal-containing PF medium in the BacT/alert blood culture system. J. Clin. Microbiol. 52: 1898–1900.
Rajapaksha, P., A. Elbourne, S. Gangadoo, R. Brown, D. Cozzolino, and J. Chapman (2019) A review of methods for the detection of pathogenic microorganisms. Analyst. 144: 396–411.
Opota, O., K. Jaton, and G. Greub (2015) Microbial diagnosis of bloodstream infection: towards molecular diagnosis directly from blood. Clin. Microbiol. Infect. 21: 323–331.
Anis, E., I. K. Hawkins, M. R. S. Ilha, M. W. Woldemeskel, J. T. Saliki, and R. P. Wilkes (2018) Evaluation of targeted next-generation sequencing for detection of bovine pathogens in clinical samples. J. Clin. Microbiol. 56: e00399–18.
Yoo, S. M., S. Y. Lee, K. H. Chang, S. Y. Yoo, N. C. Yoo, K. C. Keum, W. M. Yoo, J. M. Kim, and J. Y. Choi (2009) High-throughput identification of clinically important bacterial pathogens using DNA microarray. Mol. Cell. Probes. 23: 171–177.
Yoo, S. M., J. Y. Choi, J. K. Yun, J. K. Choi, S. Y. Shin, K. Lee, J. M. Kim, and S. Y. Lee (2010) DNA microarray-based identification of bacterial and fungal pathogens in bloodstream infections. Mol. Cell. Probes. 24: 44–52.
Tissari, P., A. Zumla, E. Tarkka, S. Mero, L. Savolainen, M. Vaara, A. Aittakorpi, S. Laakso, M. Lindfors, H. Piiparinen, M. Mäki, C. Carder, J. Huggett, and V. Gant (2010) Accurate and rapid identification of bacterial species from positive blood cultures with a DNA-based microarray platform: an observational study. Lancet. 375: 224–230.
Thanthrige-Don, N., O. Lung, T. Furukawa-Stoffer, C. Buchanan, T. Joseph, D. L. Godson, J. Gilleard, T. Alexander, and A. Ambagala (2018) A novel multiplex PCR-electronic microarray assay for rapid and simultaneous detection of bovine respiratory and enteric pathogens. J. Virol. Methods. 261: 51–62.
Yoo, S. M. and S. Y. Lee (2016) Optical biosensors for the detection of pathogenic microorganisms. Trends Biotechnol. 34: 7–25.
Knabl, L., W. Mutschlechner, and D. Orth-Höller (2016) Evaluation of a multiplex OnSpot primer-extension PCR assay in the diagnosis of sepsis. J. Microbiol. Methods. 120: 91–93.
De Angelis, G., B. Posteraro, E. De Carolis, G. Menchinelli, F. Franceschi, M. Tumbarello, G. De Pascale, T. Spanu, and M. Sanguinetti (2018) T2Bacteria magnetic resonance assay for the rapid detection of ESKAPEc pathogens directly in whole blood. J. Antimicrob. Chemother. 73: iv20–iv26.
Patch, M. E., E. Weisz, A. Cubillos, S. J. Estrada, and M. A. Pfaller (2018) Impact of rapid, culture-independent diagnosis of candidaemia and invasive candidiasis in a community health system. J. Antimicrob. Chemother. 73: iv27–iv30.
Stevenson, M., A. Pandor, M. Martyn-St James, R. Rafia, L. Uttley, J. Stevens, J. Sanderson, R. Wong, G. D. Perkins, R. McMullan, and P. Dark (2016) Sepsis: the LightCycler SeptiFast Test MGRADE®, SepsiTest™ and IRIDICA BAC BSI assay for rapidly identifying bloodstream bacteria and fungi - a systematic review and economic evaluation. Health Technol. Assess. 20: 1–246.
Pilecky, M., A. Schildberger, D. Orth-Höller, and V. Weber (2019) Pathogen enrichment from human whole blood for the diagnosis of bloodstream infection: prospects and limitations. Diagn. Microbiol. Infect. Dis. 94: 7–14.
Nguyen, Q. H. and M. I. Kim (2020) Nanomaterial-mediated paper-based biosensors for colorimetric pathogen detection. Trends Analyt. Chem. 132: 116038.
Le, T. N., T. D. Tran, and M. I. Kim (2020) A convenient colorimetric bacteria detection method utilizing chitosan-coated magnetic nanoparticles. Nanomaterials (Basel). 10: 92.
Keum, K. C., S. M. Yoo, S. Y. Lee, K. H. Chang, N. C. Yoo, W. M. Yoo, J. M. Kim, J. Y. Choi, J. S. Kim, and G. Lee (2006) DNA microarray-based detection of nosocomial pathogenic Pseudomonas aeruginosa and Acinetobacter baumannii. Mol. Cell. Probes. 20: 42–50.
Humphries, R. M., J. Ambler, S. L. Mitchell, M. Castanheira, T. Dingle, J. A. Hindler, L. Koeth, and K. Sei (2018) CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J. Clin. Microbiol. 56: e01934–17.
Wiegand, I., K. Hilpert, and R. E. W. Hancock (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3: 163–175.
Uslan, D. Z., S. J. Crane, J. M. Steckelberg, F. R. Cockerill 3rd, J. L. St Sauver, W. R. Wilson, and L. M. Baddour (2007) Age- and sex-associated trends in bloodstream infection: a population-based study in Olmsted County, Minnesota. Arch. Intern. Med. 167: 834–839.
Li, L. and H. Huang (2017) Risk factors of mortality in bloodstream infections caused by Klebsiella pneumonia: A single-center retrospective study in China. Medicine (Baltimore). 96: e7924.
Durdu, B., I. N. Hakyemez, S. Bolukcu, G. Okay, B. Gultepe, and T. Aslan (2016) Mortality markers in nosocomial Klebsiella pneumoniae bloodstream infection. Springerplus. 5: 1892.
Vardakas, K. Z., D. K. Matthaiou, M. E. Falagas, E. Antypa, A. Koteli, and E. Antoniadou (2015) Characteristics, risk factors and outcomes of carbapenem-resistant Klebsiella pneumoniae infections in the intensive care unit. J. Infect. 70: 592–599.
Schleibinger, M., C. L. Steinbach, C. Töpper, A. Kratzer, U. Liebchen, F. Kees, B. Salzberger, and M. G. Kees (2015) Protein binding characteristics and pharmacokinetics of ceftriaxone in intensive care unit patients. Br. J. Clin. Pharmacol. 80: 525–533.
Ravan, H., S. Kashanian, N. Sanadgol, A. Badoei-Dalfard, and Z. Karami (2014) Strategies for optimizing DNA hybridization on surfaces. Anal. Biochem. 444: 41–46.
Yoo, S. M., D. M. Kim, and S. Y. Lee (2015) Multispot array combined with S1 nuclease-mediated elimination of unpaired nucleotides. BioChip J. 9: 156–163.
Milton, J. A., S. Patole, H. Yin, Q. Xiao, T. Brown, and T. Melvin (2013) Efficient self-assembly of DNA-functionalized fluorophores and gold nanoparticles with DNA functionalized silicon surfaces: the effect of oligomer spacers. Nucleic Acids Res. 41: e80.
Wirtz, R., C. Wälti, P. Tosch, M. Pepper, A. G. Davies, W. A. Germishuizen, and A. P. J. Middelberg (2004) Influence of the thiol position on the attachment and subsequent hybridization of thiolated DNA on gold surfaces. Langmuir. 20: 1527–1530.
Wick, L. M., J. M. Rouillard, T. S. Whittam, E. Gulari, J. M. Tiedje, and S. A. Hashsham (2006) On-chip non-equilibrium dissociation curves and dissociation rate constants as methods to assess specificity of oligonucleotide probes. Nucleic Acids Res. 34: e26.
Yilmaz, L. S., A. Loy, E. S. Wright, M. Wagner, and D. R. Noguera (2012) Modeling formamide denaturation of probetarget hybrids for improved microarray probe design in microbial diagnostics. PLoS One. 7: e43862.
Regué, M., B. Hita, N. Piqué, L. Izquierdo, S. Merino, S. Fresno, V. J. Benedí, and J. M. Tomás (2014) A gene, uge, is essential for Klebsiella pneumoniae virulence. Infect. Immun. 72: 54–61.
Acknowledgements
This work was supported by an NRF grant funded by the Ministry of Science and ICT (NRF-2019R1A2C1088504) and the Chung-Ang University Research Scholarship Grants in 2019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shin, S.Y., Kim, D.M., Jo, Y. et al. DNA Microarray-based Detection of Bacteria in Samples Containing Antibiotics: Effect of Antibiotics on the Performance of Pathogen Detection Assays. Biotechnol Bioproc E 26, 447–455 (2021). https://doi.org/10.1007/s12257-020-0342-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-020-0342-9