Abstract
Gastric cancer (GC) is a common malignant solid tumor that is characterized by high hypoxia. The transcription of genes associated with hypoxia affects tumor occurrence and development. Long non-coding RNAs (lncRNAs) have been reported to play important roles in cancer development. In this study, we screened for differentially expressed ncRNAs (non-coding RNA) and mRNAs between hypoxia-inducible factor-1 (HIF-1α) knockdown GC cells and scrambled GC cells. Microarray data revealed that HIF-1α regulated the expression of LINC01355 (Hypoxia Yield Proliferation Associated LncRNA, HYPAL). HYPAL was found to be significantly upregulated in GC cells and tissues and was correlated with poor GC prognosis. Chromatin immunoprecipitation (ChIP) and luciferase reporter assays revealed that HIF-1α promoted HYPAL expression by binding the promoter region. A regulatory network for the competing endogenous RNA (ceRNA) was constructed using bioinformatics tools. Mechanistic studies revealed that HYPAL acted as a ceRNA of miR-431-5p to regulate CDK14 expression. Carcinogenic effects of HYPAL were evaluated in vitro and in vivo. The HIF-1α/HYPAL/miR-431-5p/CDK14 (Cyclin-dependent kinase 14) axis activated the Wnt/β-catenin signaling pathway and induced GC cell proliferation while inhibiting apoptosis. In conclusion, HYPAL is a potential molecular target for GC therapy.
Similar content being viewed by others
Background
Gastric cancer (GC) is the most common malignant tumor of the digestive system. Despite advances in therapy, GC remains the second leading cause of cancer-related mortalities in Asia [1]. This outcome is associated with its high malignant proliferative and invasive rates [2]. The tumor microenvironment provides necessary factors for tumor cell proliferation and plays an important role in tumor growth as well as diffusion [3]. In most solid tumors, hypoxia is a significant feature of the microenvironment and has been shown to play a crucial role in cell proliferation, angiogenesis, chemoresistance, and invasion [4]. In our previous studies, we established the roles of hypoxia and hypoxia-inducible factor-1 (HIF-1α) as onco-factors in GC occurrence and development and revealed that they were markers for poor prognosis [5, 6]. Moreover, we found that remodeling of the hypoxic microenvironment in GC might be associated with impaired expression of long non-coding RNAs (lncRNAs) [7, 8].
LncRNAs are highly heterogeneous RNA molecules, about 200 nucleotides in length, with a limited protein-coding ability [9]. LncRNAs are widely found in introns, intergene DNA, or antisense gene overlap, while some of them can only be detected at specific developmental stages or tissues. Structurally and functionally, lncRNAs are highly similar to messenger RNA (mRNAs) with regards to polyadenylation, splicing, capping, and RNA polymerase II-based transcription [10]. Various studies have evaluated the association between lncRNAs and GC, and revealed the underlying mechanisms of GC pathogenesis and progression [11,12,13]. Notably, hypoxia-responsive lncRNAs (HRLs), a specific group of lncRNAs that are closely associated with hypoxia, have been described. The HRLs regulate the HIFs pathway through two main mechanisms [14, 15]. First, HRLs could regulate the HIFs pathway directly. Second, HIFs bind the hypoxia-responsive elements in the promoter region of HRLs or regulate HRLs by inducing the methylation of its promoter or through the molecular sponge action of microRNAs (miRNAs). However, the specific patterns of HRLs, as well as their roles in GC malignant proliferation and invasion, have not been clearly established.
In this study, we aimed to evaluate HRLs expression levels under hypoxia conditions and elucidate their effects on cell proliferation. Specifically, we knocked down HIF-1α in GC cells and analyzed the corresponding changes in transcriptomics. We found that LINC01355 (NR_110616) was overexpressed in GC cells and tissues, and was closely associated with poor prognosis of GC. Next, we investigated the role of LINC01355 in cell proliferation and cell cycle, as well as apoptosis in GC. Our results showed that loss of function of transcription factor HIF-1α significantly downregulated LINC01355, while HIF-1α could bind to the promoter region of LINC01355 to induce LINC01355 transcription. Furthermore, LINC01355 could activate the Wnt/β-catenin signaling pathway through competitive endogenous RNAs (ceRNA) mechanism to induce malignant proliferation of GC. Thus, we termed LINC01355 as Hypoxia Yield Proliferation Associated LncRNA (HYPAL). Our work shed light on an original signaling axis in the hypoxia microenvironment and provides a potential therapeutic strategy for GC treatment.
Materials and methods
Cell culture
The human normal gastric epithelial cell line (GES-1), GC cell line (SGC-7901, BGC-823, AGS, and MKN-45), and human embryonic kidney cell line (HEK-293) were obtained from China Medical University (Shenyang, China). Cells were maintained in RPMI-1640 (or DMEM) supplemented with 10% fetal bovine serum under 1% O2 or 20% O2 conditions as previously described [7, 8].
Cell transfection and establishment of stable lines
SGC-7901 and BGC-823 cells were plated into 6-well plates, at 80% confluence, they were transfected with lentiviruses harboring shRNA sequences targeting human HIF-1α, negative control shRNA (sh-NC, GeneChem, China), and LINC01355 shRNAs, CDK14 shRNAs (Invitrogen, USA). LINC01355 and CDK14 expression vectors were constructed using the pcDNA3.1 backbone vector (Invitrogen, USA). The miR-431-5p mimic, miR-431-5p inhibitor, miRNA negative control (miR-NC) were purchased from Invitrogen, USA. Cell transfections were performed using the Lipofectamine® 2000 Reagent (Invitrogen, USA) according to the manufacturer's instructions, and as previously described [8].
We transfected the expression vector (1 μg), shRNA (50 nM), or miRNA (50 nM) into 108 of SGC-7901 and BGC-823 cells, with untransfected cells also included as controls. For NC experiments, we transfected a similar number of cells with the same amount of empty expression vector, NC shRNAs or pcDNA3.1, and cultured them in a fresh medium for 48 h. Sequences for shRNAs, miRNAs were listed in Supplementary Table 1.
Recombinant CDK14 lentiviruses and sh-LINC01355 lentiviruses were transfected into GC cell lines, respectively, according to the manufacturer’s instructions. Then, cells were treated with 2 mg/L puromycin (Invivogen, USA) for two weeks to obtain cells with stable overexpression of CDK14 or LINC01355 knockdown.
Experimental tissues and ethical approval
GC tissues and the adjacent non-cancerous tissue samples were obtained from 50 patients who had been subjected to surgical resection at Liaoning Province Cancer Hospital and Institute. Sample collection was performed between 2014 and 2015. Patient follow-up was done up to December 31, 2020. Prior to enrollment in this study, patients had not been administered with either chemotherapy or radiotherapy. Before recruitment, all patients were required to sign a written informed consent. Ethical approval was obtained from the Ethical Committee of Liaoning Province Cancer Hospital and Institute (20181226).
Microarray analysis and ceRNA prediction
Total RNA was extracted from SGC-7901 and HIF-1α knockdown SGC-7901 cells, amplified, and transcribed into cRNA using the Quick Amp Labeling kit (Agilent Technologies, USA). Labeled cRNAs were hybridized to Human LncRNA Array v3.0 (8 × 60 K, ArrayStar, USA) using Agilent Technologies. Then, the miRCURYTM (Hy3TM/Hy5TM) power labeling kit (Exiqon Life Sciences, Denmark) was used to label the total miRNAs. The labeled miRNAs were hybridized onto the miRCURY LNA microRNA array (v18.0, Exiqon Life Sciences), washed, and scanned using the Axon GenePix 4000B microarray scanner. Array images were extracted and analyzed using an Agilent Feature Extraction Software V10.7. Next, we used the GeneSpring GX V11.5.1 software (Agilent Technologies) to normalize the original signal strength and to filter low-strength lncRNAs, mRNAs, and miRNAs. Upregulated or downregulated lncRNAs, mRNAs, and miRNAs were detected using fold change > 2.0 and p < 0.05 as the thresholds (Supplement Data 1).
With LINC01355 as the core molecule, miRanda (a dynamic programming algorithm for secondary structure and free energy) and TargetScan (for site prediction and conservative scoring) algorithms were used for target prediction. The miRNA information in the latest miRBase was used in the process. The parameters were: miRNAcoverage > 0.3, context + < 0.05 (TargetScan v6.0), context < 0.05 (TargetScan v5.0), Energy < − 10 (miRanda), structure > 140 (miRanda), P < 0.05. Finally, the Cytoscape 3.6.1 software (http://www.cytoscape.org/) was used to construct and visualize the ceRNA network.
Expression analysis
Total RNAs from GC tissues, the adjacent non-cancerous tissues, and from GC cells were extracted using the TRIZOL kit (Invitrogen, USA) according to the manufacturer’s instructions. Total RNAs were reverse-transcribed into complementary deoxyribose nucleic acid (cDNA) using Revert aid first-strand cDNA synthesis kit (Thermo Fisher, USA). Then, samples were subjected to quantitative real time polymerase chain reaction (qRT-PCR) using the SYBR Green One-Step RT-PCR Master Mix (Thermo Fisher, USA). Amplification was performed on a 7900 real-time PCR system (Applied Biosystems, USA), as previously described [8]. Primer sequences of target genes were designed by Sangon (Supplementary Table 1).
Separation of nuclear and cytoplasmic fractions
Total cellular fractions were divided into nuclear and cytoplasmic fractions using the PARIS kit (AM1921; Thermo Fisher Scientific, Japan) according to the manufacturer’s instructions and as previously described [8]. The kit was used to isolate RNA from same experimental samples and ensures that nuclear and cytoplasmic components were isolated prior to RNA isolation.
Fluorescence in situ hybridization (FISH)
A LINC01355 probe was designed and synthesized by RiboBio (Guang Zhou, China). Cells were cultured overnight in a 6-well plate, fixed in 4% paraformaldehyde for 30 min, and treated with 0.5% Triton X-100 for 20 min. Cells were washed using 2 × Buffer for 10 min, after which they were incubated with the LINC01355 probe. Slides were fixed with an extended gold anti-fading agent and tested with 4′, 6-diamino-2-phenylindoles (DAPI, Invitrogen, USA). LINC01355 was observed under a Leica-SP8 confocal microscope (Leica, Germany).
Chromatin immunoprecipitation (ChIP)
ChIP detection was performed using the simple ChIP enzymatic chromatin IP kit protocol (Cell Signaling Technology, China) [16]. Briefly, GC cells were cultured in the medium for 48 h, and 1 × 107 cells subjected to ChIP. Chromatin was immunoprecipitated for 16 h at 4 °C on rotators with an anti-HIF-1α (Santa Cruz Biotechnology, USA, 1:50) and mouse IgG (Santa Cruz Biotechnology, USA, 1:100). Purified LINC01355 DNA fragments, targeting the LINC01355 promoter, were used as templates for PCR analysis.
Dual-luciferase, promoter-luciferase, and TOPFlash/FOPFlash reporter assays
We performed a dual-luciferase reporter assay to determine whether miR-431-5p bound the 3'UTR of LINC01355 and CDK14 (Promega, USA). Briefly, we constructed psiCHECK2 vector (Promega, USA) reporter plasmids harboring wild-type (wt) and mutant-type (mut) LINC01355 and CDK14 3′-UTR sequences with predicted binding sites for miR-431-5p. The vector was transfected using the POLO3000 transfection reagent (Research and science, China) while HEK-293 cells were co-transfected with reporter plasmids and miR-431-5p mimics or miR-NC.
The promoter-luciferase reporter assay was performed to detect whether HIF-1α could bind the LINC01355 promoter. The recombinant pGL-3 basic-plasmid contained truncated human LINC01355 promoter (wild type, wt) or mutant LINC01355 promoters (mutant type, mut). Whole wt and mut sequences are shown in Supplement Data 2. The promoter-reporter plasmid and HIF-1α (or NC) vectors were co-transfected using the POLO3000 transfection reagent (Research and science, China). Forty-eight hours later, luciferase activity was evaluated in each group according to the manufacturer’s instructions.
For the TOPFlash/FOPFlash reporter assay, Wnt/β-catenin signaling reporter TOPFlash/FOPFlash (Upstate Biotechnology, USA) was co-transfected into cells along with HYPAL vectors, CDK14 vectors, or miR-431-5p mimics. Experiments were performed in both HEK-293 and SGC-7901, and repeated three times, respectively.
Western blot analysis
Total proteins were extracted from SGC-7901 and BGC-823 cells using the RIPA lysis buffer (Beyotime, Shanghai, China) and stored at − 80 °C until use. Total proteins were separated on a 10% SDS-PAGE and transferred onto PVDF membranes. After blocking, treated membranes were incubated overnight with primary antibodies (CDK14, 1:1000, Santa, sc-376366; β-Catenin, 1:1000, CST, 9582S; phoshpo-β-Catenin (Ser33/37/Thr41), 1:1000, CST, 9561; Survivin, 1:5000, Abcam, ab76424; c-myc, 1:1000, Abcam, ab32072; MMP-9, 1:1000, CST, 2270S) at 4℃, then with the secondary antibody for 2 h. Finally, protein bands were visualized on an ECL system (Amersham Biosciences, UK).
Immunohistochemistry
Immunohistochemical analysis was performed as previously described [6]. Ki67 (1:1000, Abcam, ab15580), Caspase-3 (1:1000, Abcam, ab184787).
Colony formation assay
After transfection, a total of 1 × 103 SGC-7901 and BGC-823 cells were seeded into 6-well plates and incubated for 2 weeks. The resulting cell colonies were washed three times using PBS, fixed with 4% paraformaldehyde, and stained with 0.1% crystal violet solution (Sigma). The number of stained colonies was observed using a light microscope.
Cell counting assay
Cell counting was performed using the cell counting kit (CCK-8), according to the manufacturer’s instructions. Briefly, cells in each group, at the logarithmic growth stage, were inoculated into 96 well plates, and incubated at 37 ℃ for 24, 48, 72, and 96 h. Then, the medium was discarded and a chromogenic solution prepared at a ratio of 10:1. Thereafter, 10 μl of color developing solution was added into the 96 well plates, followed by 2 h of incubation at 37 ℃. Optical densities (OD) for each group were detected using an ultraviolet spectrophotometer at 450 nm.
Proliferation assay
Proliferations of SGC-7901 and BGC-823 cells were assessed using a 5-ethynyl-20-deoxyuridine (EdU) assay kit (RiboBio, Guangzhou, China). Briefly, transfected SGC-7901 and BGC-823 cells were incubated with 50 μm EdU for 2 h after which cell nuclei were dyed with 4′, 6-diamidino-2phenylindole (DAPI, Sigma, USA). Finally, EdU-positive cells were observed under a fluorescence microscope.
Cell apoptosis assay
The apoptosis of SGC-7901 and BGC-823 cells were assessed using an AnnexinV-fluorescein isothiocyanate/propidium iodide (FITC/PI) apoptosis detection kit (Invitrogen, USA) according to the manufacturer’s instructions. Summarily, transfected SGC-7901 and BGC-823 cells were harvested and resuspended in a binding buffer, then stained with Annexin V-FITC and PI. Apoptotic cells were examined using a flow cytometer (BD Biosciences, USA).
Cell cycle analysis
Cells were resuspended in pre-chilled 1 × PBS, then diluted to 1 × 105 cells/ml using the 1 × Annexin binding buffer. Cells were precooled using 70% ethanol at 4 °C overnight before assaying.
Animal experiments
Approximately 1 × 107 of stably transfected SGC-7901 cells were resuspended in 0.2 mL PBS, and subcutaneously injected into the right axillary region of female five-week-old BALB/ C nude mice. Then, tumor sizes were measured every 3 days using vernier calipers. Their lengths (L) and widths (W) were recorded and used to calculate their volume using the formula: Tumor volume = L*W*W/2. Twenty one days after injection, mice were euthanized, their subcutaneous tumors isolated and weighted.
Statistical analysis
Data were analyzed using GraphPad Prism 9.0 (GraphPad Software Inc) and presented as means ± standard deviations (SD). Statistical comparisons between two groups were performed using the Student’s t-test, while comparisons among groups were performed using one-way analysis of variance (ANOVA) followed by Tukey's post hoc test. P < 0.05 was considered statistically significant.
Results
Hypoxia responsive lncRNAs HYPAL is overexpressed in GC cells
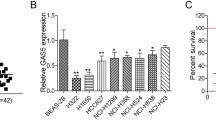
To determine the RNA that is associated with hypoxia and to analyze the regulatory network of ncRNA and mRNA in GC, we knocked down HIF-1α expression in SGC-7901 (sh-HIF-1α) and used scrambled GC cells as controls (Crtl). Our findings were in accordance with our previous study, which showed that sh-HIF-1α#2 exhibited the best silencing efficiency (Fig. S1A and B) [8]. We used the Arraystar Human lncRNA + mRNA Microarray V4.0 and Whole Human miRNA Microarray platforms to detect differentially expressed RNAs between sh-HIF-1α and Ctrl. A total of 311 differentially expressed lncRNAs were detected, among which 133 and 178 were, respectively, upregulated and downregulated. A total of 507 miRNAs were significantly differentially expressed, among which 286 and 221 were upregulated and downregulated, respectively. Moreover, 48 mRNAs (with 25 and 23 upregulated and downregulated, respectively) were differentially expressed (Supplement Data 1, P < 0.05, Fold change > 2). We used a hierarchical clustering heatmap to show differences among target lncRNAs, miRNAs, and mRNAs (Fig. 1a). The depletion of transcription factor HIF-1α inhibited gene transcription and downregulated the expression of corresponding lncRNAs. Therefore, we focused on the downregulated lncRNAs in sh-HIF-1α. Sixty-six (66) of the 178 downregulated lncRNAs were intergenic lncRNAs, while 15 were confirmed in GENCODE and RefSeq databases. We combined our findings with results from The Cancer Genome Atlas (TCGA) database. Among them, 7 lncRNAs were overexpressed in the transcriptome data of TCGA-GC (STAD). Subsequently, we performed qRT-PCR verification on the 7 potential onco-lncRNAs. We found that LINC01355 (NR_110616, HYPAL) was overexpressed in both GC tissues and cells (Fig. 1b–d), implying that it might be a potential carcinogenic factor. Moreover, HYPAL levels were elevated in GC cells under hypoxic conditions (1% O2, Fig. 1e), while nuclear/cytoplasmic RNA fractionation and immunofluorescence assays indicated that it was mainly localized in the cytoplasms of both SGC-7901 and BGC-823 cells (Fig. 1f, g). These findings imply that HYPAL might exhibit its functions at the post-transcription level.
HYPAL was a hypoxia associated lncRNA and overexpressed in GC. a Hierarchical clustering analysis of lncRNAs and miRNAs, and mRNAs that were differentially expressed between HIF-1α knockdown GC cells and control GC cells (p < 0.05, fold change > 2; filtered to show the top 25 upregulated or downregulated results for RNAs). Expression values were represented in shades of red and green, indicating expression above and below the median expression value across all tissues, respectively. The red arrow pointed to HYPAL. b HYPAL was overexpressed in GC tissue in TCGA. c HYPAL was overexpressed in clinical GC tissue sample. d HYPAL was upregulated in GC cells. e HYPAL was upregulated under hypoxic culture conditions in GC cells. f, g The expression level of HYPAL in the subcellular fractions of SGC-7901 and BGC-823 cells was detected by qRT-PCR. U6 and β-actin were used as nuclear and cytoplasmic markers, respectively. h Immunofluorescence indicates that HYPAL was mainly localized in the cytoplasm (U6 and β-actin were used as a loading control in qRT-PCR; data are shown as mean ± SD, n = 3. The data statistical significance is assessed by Student’s t test. *p < 0.05, **p < 0.01)
HYPAL is associated with poor survival of GC patients
We used the clinical data of the 50 patients to assess the clinical significance of HYPAL in GC. Disease-free survival (DFS) and overall survival (OS) outcomes ranged from 5 to 85 and 18–85 months, respectively, while 40 (80%) patients died before the end of the follow-up period. The correlation between HYPAL expression and other clinic-pathological factors revealed a significant correlation between its overexpression with TNM stage (Table 1). Furthermore, HYPAL, Bormann type, tumor size, and TNM stages were closely correlated with DFS and OS, and they indicated a poor prognosis (Fig. 2a, b, Table 2). Besides, HYPAL was found to be an independent risk factor for poor survival outcomes of GC patients (Table 3).
HIF-1α affected the expression of HYPAL directly. a, b Kaplan–Meier analysis of the correlation between HYPAL expression levels and disease-free survival and overall survival. c HIF-1α shRNA inhibited the expression of HYPAL; d Luciferase assays were performed in 293A cells transfected with wt or mut promoter. e The interaction of HIF-1α with HYPAL was shown using ChIP assays with control (IgG) or anti-HIF-1α antibody (data are shown as mean ± SD, n = 3. The data statistical significance is assessed by Student’s t test. *p < 0.05, **p < 0.01)
HIF-1α activates HYPAL in GC Cells
Microarrays revealed suppressed HYPAL expressions in sh-HIF-1α GC. To determine whether HIF-1α regulated HYPAL expression, we transfected SGC7901 and BGC-823 cell lines with sh-HIF-1α#2, which had the best silencing efficiency (Fig. S1). Transfection with sh-HIF-1α#2 downregulated HYPAL in both GC lines (Fig. 2c), implying that HIF-1α regulates HYPAL expression. To confirm whether HIF-1α could directly bind the HYPAL promoter in vitro, we cloned a full-length HYPAL promoter into a luciferase reporter plasmid. The dual-luciferase reporter assay revealed significantly elevated fluorescence intensities of OE-HIF-1α in cells treated with HYPAL-wt relative to those treated with OE-NC. However, there were no significant difference in fluorescence intensities after transfections of OE-NC and OE-HIF-1α with HYPAL-mut (Fig. 2d). Subsequently, ChIP analysis revealed that the HYPAL promoter was pulled down by the HIF-1α antibody but not by the control IgG (Fig. 2e). Figure 2f showed the binding site. These findings confirm that HYPAL overexpression under a hypoxic environment was due to the direct actions of transcription factor HIF-1α. In other words, HIF-1α promoted HYPAL expression. The full-length sequence (HYPAL-wt, HYPAL-mut) was listed in Supplementary Data 2.
HYPAL can crosstalk with miR-431-5p through direct binding
To evaluate the biological functions of HYPAL and verify its association with miRNAs and mRNAs, we performed HYPAL-ceRNA analysis using microarray data for lncRNAs, miRNAs, and mRNAs (Fig. 3a). miR-431-5p, miR-27-3P, miR-145-5p, and miR-888-5p were found to be potential HYPAL targets. Subsequently, we verified the effect of HYPAL sh-RNAs and pcDNA3.1-HYPAL on the expression of HYPAL and found that the first sequence exhibited a better silencing efficiency (Fig. S2a, b). Inhibition of HYPAL upregulated miR-431-5p. However, it did not significantly alter the expression of the other three miRNAs (Fig. 3b). Similarly, miR-431-5p mimics suppressed HYPAL expression (Fig. 3c), whereas miR-431-5p was downregulated in GC cells and tissues (Fig. 3d, e). Spearman correlation analysis revealed that HYPAL was negatively correlated with miR-431-5p expression (Fig. 3f, r = − 0.72, p < 0.01). The inhibitor and mimics reversed miR-431-5p expression patterns (Fig. S2c). Moreover, we used DianaTools (mihttp://diana.imis.athena-innovation.gr/DianaTools) to predict the potential binding sites for HYPAL’s 3’UTR and miR-431-5p (Fig. 3g). The overexpression of miR-431-5p-wt, but not miR-431-5p-mut, or miRNA negative control (miR-NC) suppressed the luciferase activity of psiCHECK2-HYPAL (Fig. 3h).
HYPAL was identified as a target of miR-431-5p. a The lncRNA (HYPAL)-miRNA-mRNA networks in GC. b The knockdown of HYPAL promoted the expression of miR-431-5p. c The mimics of miR-431-5p inhibited the expression of HYPAL. d, e miR-431-5p was weak-expressed in GC cells and tissues. f The expressions of HYPAL and miR-431-5p were negatively correlated. g Predicted binding sites for miR-431-5p on the HYPAL transcript. The white nucleotides are the seed sequences of miRNAs. h Luciferase activity in HEK-293 cell co-transfected with luciferase reporter containing HYPAL and the mimics of miR-431-5p or mutant. Data are presented as the relative ratio of renilla luciferase activity and firefly luciferase activity (data are shown as mean ± SD, n = 3. The data statistical significance are assessed by Student’s t test. *p < 0.05, **p < 0.01)
The biological function of HYPAL in vitro
We evaluated the roles of HYPAL and miR-431-5p in GC by downregulating HYPAL in hypoxic-cultured and overexpressing HYPAL in normoxia-cultured GC cells, then evaluating the resultant effects on cell proliferation, apoptosis, and cell cycle.
The CCK-8 assay showed that transfecting sh-HYPAL into GC cells inhibited cell proliferation under hypoxia conditions. Simultaneously, suppressing miR-145-5p expression ameliorated this tumor suppressor effect (Fig. 4a, b). Upregulated HYPAL significantly enhanced cell proliferation under normoxia, while miR-431-5p mimics rescued the phenotype (Fig. S3a, b). In addition, we confirmed that the miR-431-5p inhibitor promoted GC proliferation, and the overexpression of miR-431-5p suppressed the proliferation (Fig. S2d). Colony formation and EdU assays revealed that downregulating HYPAL significantly suppressed clone formation and fluorescence intensities in hypoxia GC cells. However, these phenotypes were recovered after inhibiting miR-431-5p expression (Fig. 4c, d). Moreover, HYPAL overexpression increased the number of cell clones and enhanced fluorescence, although upregulated miR-431-5p counteracted this carcinogenic effect under normoxia conditions (Fig. S3c, d).
HYPAL reinforced the proliferative capacity of hypoxia GC cells. a, b The inhibitor of miR-431-5p could reverse the inhibitory effect of sh-HYPAL on hypoxia culture GC cell proliferation which were determined CCK-8 assay. c, d Cell proliferation assessed in HYPAL knockdown and HYPAL knockdown + miR-431-5p inhibitor GC cells by colony formation and EdU assay. e, f The cell cycle progression of GC cells transfected with sh-NC, sh-HYPAL, sh- HYPAL + miR-431-5p inhibitor was identified by flow cytometry assay. g, h Effects of sh-HYPAL, sh-HYPAL + miR-431-5p inhibitor on GC cell apoptosis (data are shown as mean ± SD, n = 3. The data statistical significance are assessed by Student’s t test. *p < 0.05)
Flow cytometry assay showed that sh-HYPAL significantly increased the apoptotic rate if GC cells in hypoxia, while downregulating miR-431-5p enhanced apoptosis (Fig. 4g, h). Conversely, overexpressing HYPAL inhibited apoptosis while overexpressing miR-431-5p weakened this phenotype in normoxia cells (Fig. S3g, h). The loss of HYPAL function was shown to upregulate a proportion of the S phase while downregulating the G2/M phases in hypoxia GC cells. Moreover, addition of the miR-431-5p inhibitor weakened sh-HYPAL's inhibitory effect on the cell cycle (Fig. 4e, f). Overexpressing HYPAL significantly downregulated S phase normoxia GC cells but upregulated the G2/M phase ratio. Furthermore, the miR-431-5p mimics weakened the effect of OE-HYPAL on the cell cycle (Fig. S3e, f).
Consequently, we selected HYPAL and its related regulatory ceRNA networks for further analyses. qRT-PCR and luciferase assays confirmed that HYPAL is a target molecule of miR-431-5p, and further revealed interactions between the two molecules during malignant GC proliferation.
HYPAL functioned as ceRNA of CDK14 by sponging miR-431-5p
We identified mRNAs that potentially interact with miR-431-5p and serve as ceRNAs by performing KEGG pathway analyses on the weak-expressed mRNAs in the sh-HIF-1α group (Supplement Data 1). We focused on differentially expressed genes that might be involved in tumor proliferation as well as on tight junction-related genes (Fig. 5a). CDK14 is a newly discovered member of the Cyclin-dependent kinases (CDKs) family, and it can regulate cell cycle progression by interacting with cyclins [17]. Findings from KEGG, ceRNAs network, and miR-431-5p target gene prediction revealed that CDK14 might be an important downstream target. TargetScan (http://www.targetscan.org) revealed potential sites where CDK14 and miR-431-5p could directly bind (Fig. 5b). First, we used qRT-PCR and western-blot analyses to detect CDK14 expression at mRNA and protein levels, respectively, in GC cells (Fig. 5c, d), and established its overexpression in GC relative to normal tissues (Fig. 5e–g). The siRNAs and overexpressed vector modulated CDK14 expression (Fig. S2e, f). Moreover, overexpressing miR-431-5p downregulated CDK14 at both transcription and post-transcriptional levels (Fig. 5h, i). We found a significant negative correlation between CDK14 and miR-431-5p expression in GC tissues (Fig. 5j, r = − 0.71, p < 0.01). The subsequent luciferase assay confirmed that miR-431-5p can directly bind the 3′-UTR of CDK14 (Fig. 5k) while inhibition of HYPAL downregulated CDK14 at both mRNA and protein levels (Fig. 5l, m). These results indicate that HYPAL, as a ceRNA, competitively binds miR-431-5p, thereby affecting CDK14 expression.
CDK14 could crosstalk with miR-431-5p through direct binding. a Differentially expressed genes were enriched in the tight junction pathway. b Predicted binding sites for miR-431-5p on the CDK14 transcript. c, d CDK14 was overexpressed in GC cells compare to it in GES-1 by qRT-PCR and western-blot. e, f CDK14 was overexpressed in GC tissue. g Representative images of CDK14 in GC tissue (up). CDK14 was strongly expressed in GC tissue; but weak-expressed in adjacent non-cancerous tissue samples (low, magnifications were 200 ×). h, i GC cells were transfected with the mimics of miR-431-5p which reduced CDK14 expression was shown by qRT-RCR and western-blot. j The expressions of CDK14 and miR-431-5p were negatively correlated. k Luciferase activities were measured in HEK-293 cells co-transfected with luciferase reporter containing CDK14 and the mimics of miR-431-5p or mutant. Data are presented as the relative ratio of renilla luciferase activity and firefly luciferase activity. l, m The inhibition of HYPAL could weaken the CDK14 expression at both mRNA and protein levels (data are shown as mean ± SD, n = 3. The data statistical significance is assessed by Student’s t test. *p < 0.05, **p < 0.01)
The biological function of HYPAL/miR-431-5p/CDK14 axis in vitro
We performed gain- and loss-of-function assays to understand the biological functions and regulatory relationship of CDK14 and miR-431-5p in GC cancer cells. Just as with HYPAL, we downregulated and upregulated CDK14 in hypoxic- and normoxia-cultured GC cells. The CCK-8 assay showed that downregulating CDK14 inhibited the proliferative abilities of GC cells under hypoxia conditions. Treatment of the cells with an miR-431-5p inhibitor reversed these effects (Fig. 6a, b). Under normoxia conditions, overexpressing CDK14 induced cell proliferation, although miR-431-5p mimics suppressed this effect (Fig. S4a, b). We validated this result using colony formation and EdU assays. Suppressed CDK14 expression reduced the number of cell clones as well as fluorescence intensities. However, the inhibition of miR-431-5p could enhanced cell clone formation and fluorescence intensities (Fig. 6c, d). Overexpressing CDK14 in normoxia GC cells significantly increased the number of cell clones and elevated their fluorescence intensities. However, simultaneously upregulating CDK14 and miR-431-5p significantly reduced the carcinogenic effect (Fig. S4c, d).
CDK14 contributed to the proliferation of hypoxia GC cells. a, b The inhibitor of miR-431-5p could reverse the inhibitory effect of si-CDK14 on hypoxia culture GC cell proliferation which were determined CCK-8 assay. c, d Cell proliferation assessed in CDK14 knockdown and CDK14 knockdown + miR-431-5p inhibitor GC cells by colony formation and EdU assay. e, f The cell cycle progression of GC cells transfected with si-NC, si-CDK14, si- CDK14 + miR-431-5p inhibitor was identified by flow cytometry assay. g, h Effects of si-CDK14, si-CDK14 + miR-431-5p inhibitor on GC cell apoptosis (data are shown as mean ± SD, n = 3. The data statistical significance are assessed by Student’s t test. *p < 0.05)
Downregulated CDK14 levels resulted in elevated apoptosis levels, although an opposite effect was observed when miR-431-5p was downregulated, in a similar fashion to HYPAL (Fig. 6g, h). In contrast, overexpressed CDK14 inhibited apoptosis, while miR-431-5p mimics antagonized this phenotype (Fig. S4g, h). The analysis of cell cycle progression showed that the cell cycle-related protein, si-CDK14, significantly enhanced the proportion of GC cells in the S phase but decreased their proportions in the G2/M phase. However, this phenotype could be recovered by an miR-431-5p inhibitor (Fig. 6e, f). Besides, overexpressing CDK14 significantly downregulated S phase in normoxia GC cells and upregulated the G2/M phase ratio. Consequently, exposure to miR-431-5p mimics suppressed the effect of OE-CDK14 on the cell cycle (Fig. S4e, f). These results indicate that existence of the HYPAL/miR-431-5p/CDK14 axis is necessary for HYPAL’s involvement in GC proliferation and apoptosis.
HYPAL activates the Wnt/β-Catenin pathway to induce cell proliferation
CDK14 affects cell proliferation and the cell cycle by activating the Wnt/β-catenin signaling pathway [18, 19]. Therefore, investigated the effect of HYPAL/miR-431-5p/CDK14 axis on the Wnt/β-Catenin pathway. We divided our experiment into nine groups; untreated GC cells (NC), GC cells with miR-431-5p mimics added (miR-431-5p-mimics), GC cells with mimics-NC added (mimics-NC), GC cells with overexpressed CDK14 (OE-CDK14), GC cells with control-plasmid added (NC-CDK14), GC cells with overexpressions of both miR-431-5p and CDK14, GC cells with inhibited HYPAL (sh-HYPAL), GC cells with sh-NC (sh-NC) and GC cells with inhibited HYPAL and overexpressed CDK14 (sh-HYPAL + OE-CDK14). First, we evaluated the effects of the aforementioned factors on CDK14 expression. As earlier mentioned, miR-431-5p-mimics and sh-HYPAL downregulated CDK14 expression. miR-431-5p mimics and sh-HYPAL also downregulated CDK14, even after exogenous CDK14 overexpression (Fig. 7a, S5a). Second, we detected the wnt/β-Catenin pathway-associated proteins, namely C-myc, MMP9, β-Catenin, and Survivin in the same groups and obtained comparable results to those of CDK14. Notably, overexpressed CDK14 upregulated C-myc, MMP9, β-Catenin, and Survivin to activate the Wnt/β-catenin pathway. miR-431-5p-mimics and sh-HYPAL inhibited activation of the Wnt/β-catenin pathway. Moreover, the miR-431-5p-mimics and sh-HYPAL also antagonized CDK14-triggered activation of the Wnt/β-catenin signaling pathway (Fig. 7a, the statistics data in Fig. S5b–e). As total β-catenin increased, expression levels of phosphorylated β-catenin proteins decreased (Fig. 7a, Fig. S5f). Therefore, the HYPAL/miR-431-5p/CDK14 axis inhibited β-catenin phosphorylation and its degradation so as to promote activation of the wnt/β-Catenin pathway.
In vivo tumor lumps of xenograft mouse models composed of HYPAL, miR-431-5p and CDK14. a Western blot analysis of indicated proteins confirmed that HYPAL/miR-431-5p/CDK14 axis activated the Wnt/β-catenin pathway. b, c The effect on TOP/FOP reporter activity in HEK-293 and SGC-7901 cells after transfection with control, miR-431-5p mimic or HYPAL overexpression plasmid was validated by dual-luciferase assay. d, e The effect on TOP/FOP reporter activity in HEK-293 and SGC-7901 cells after transfection with control, miR-431-5p mimic or CDK14 overexpression plasmid was validated by dual-luciferase assay. f Representative Images of the tumor lumps of each group at the endpoint of the experiment. g The tumor growth curves of in vivo tumor volumes. h. The mean tumor weight of each group. i H&E staining and immunohistochemistry of Ki67, and caspase3. Scale bar = 100 μm. j The mechanism graph of the regulatory network and function of HYPAL. HIF-1α upregulated the expression of HYPAL. The overexpressed HYPAL could promote proliferation and inhibited apoptosis processes of GC, which could be inhibited by miR-431-5p and enhanced by CDK14 as ceRNA. The HYPAL/miR-431-5p/CDK14 axis finally realized this molecular phenotype through Wnt/β-catenin pathway (data are shown as mean ± SD, n = 5. The data statistical significance is assessed by Student's t test. *p < 0.05)
We also determined whether HYPAL/miR-431-5p/CDK14 affects Wnt/β-catenin pathway activation. TOPFlash and FOPFlash reporters, containing wt and mut TCF-4 (T cell transcription factor 4) consensus binding sites, respectively, were constructed to determine whether HYPAL/miR-431-5p modulated the canonical Wnt/β-catenin pathway by regulating CDK14. The transcriptional activity of TOP/FOP was significantly enhanced with stable overexpressions of HYPAL. Overexpressed miR-431-5p reversed the increased transcriptional activity of TOP/FOP caused by OE-HYPAL in both HEK-293 and SGC-7910 cells (Fig. 7b, c). Comparable findings were observed in miR-431-5p and CDK14. The upregulated CDK14 enhanced TOP/FOP transcriptional activity. However, miR-431-5p mimics inhibited the transcriptional activity (Fig. 7d, e).
These findings imply that the HYPAL/miR-431-5p/CDK14 axis promoted cell proliferation, inhibits apoptosis and regulates cell cycle through the Wnt/β-catenin signaling pathway.
Biological functions of the HYPAL/miR-431-5p/CDK14 axis in vivo
To evaluate the biological functions of the HYPAL/miR-431-5p/ CDK14 axis in vivo, we subcutaneously injected different SGC-7901 cells into nude mice. The animals were allocated into six groups (n = 5 per group); control sh-RNA-treated GC cells (sh-NC), GC cells simultaneously overexpressing miR-431-5p and CDK14 (miR-431-5p mimics + OE-CDK14), GC cells with inhibited HYPAL (sh-HYPAL), GC cells overexpressing miR-431-5p (miR-431-5p mimics), GC cells with inhibited HYPAL but overexpressing CDK14 (sh-HYPAL + OE-CDK14), and GC cells overexpressing CDK14 (OE-CDK14). Inhibition of HYPAL or overexpression of miR-431-5p resulted in tumor mass reduction relative to the control group (sh-NC). CDK14 overexpression groups exhibited enlarged tumor lumps. However, rescue assays performed in GC cells using OE-CDK14 showed that the inhibition of HYPAL or overexpression of miR-431-5p shrank the induced subcutaneous tumors (Fig. 7f). We measured tumor volumes from the 12th day after tumor inoculation, every 3 days, sacrificed the mice at the end of the experiment on the 21st day after inoculation, then measured tumor volumes and weights across each group. Tumor volumes in the sh-HYPAL and miR-431-5p mimic groups were small, relative to the sh-NC group. However, overexpression of CDK14 increased the tumor volumes.
At the time of sacrifice, mean tumor volumes in mice injected with miR-431-5p mimics or sh-HYPAL cells were 621.32 ± 40.00 and 599.31 ± 31.00 mm3, respectively, while that for mice injected with sh-NC cells was 1021.11 ± 58.31 mm3. Upregulated CDK14 reversed this phenotype. Tumor volumes of mice injected with miR-431-5p mimics + OE-CDK14 and sh-HYPAL + OE-CDK14 were 950.31 ± 50.22 and 980.30 ± 57.61 mm3, respectively (Fig. 7g). Similarly, miR-431-5p mimics or sh-HYPAL resulted in small tumor weights (0.71 ± 0.03 or 0.61 ± 0.09 g vs sh-NC: 1.63 ± 0.20 g). However, overexpressed CDK14 reversed this phenotype (1.49 ± 0.14 g, 1.61 ± 0.05 g, miR-431-5p mimics + OE-CDK14 and si-HYPAL + OE-CDK14, Fig. 7h). To confirm the role of the HYPAL/miR-431-5p/CDK14 axis in GC cells, we performed IHC to determine Ki67 and Caspase3 protein levels in the xenografts. miR-431-5p mimics or sh-HYPAL downregulated Ki67, but upregulated Caspase3 levels. Overexpressed CDK14 reversed this outcomes (Fig. 7i). In conclusion, hypoxia promotes HYPAL expression. Furthermore, HYPAL functions as a competitive sponge for miR-431-5p, which activates the CDK14/Wnt/β-catenin pathway, thereby promoting GC proliferation (Fig. 7j).
Discussion
Advances in High-Throughput Sequencing and the progress of molecular biology have enabled researchers to gradually unravel numerous potential molecular mechanisms underlying the occurrence and development of GC [20, 21]. Particularly, uncontrolled gene expression has been shown to be a driving factor in many cancers. Genes do not function alone but as a complex regulatory network that drives the evolution of tumors through the “Cascade Effect” [22]. Besides, the interaction between tumors and their microenvironment has also been suggested as an important factor for abnormal expression of tumor-related genes [23]. For example, the hypoxic microenvironment is shown to have a non-negligible effect on the biological behavior, and malignant phenotype of GC, involving pathological angiogenesis, malignant proliferation, tumor stemness, metabolic reprogramming, invasion, and metastasis, and collaboratively promotes occurrence of GC and treatment resistance [23]. In our previous studies, we found that HIF-1α, the core element of the hypoxic microenvironment, was not only an independent risk factor for poor prognosis of GC [6], but also involved in Epithelial-Mesenchymal Transition of GC cells [5]. We also demonstrated that abnormal expression of HIF-1α could affect the expression of lncRNAs [8].
In this study, we found that regulating HIF-1α upregulated HYPAL (LINC01355 or NR_110616) expression and promoted cell proliferation. LINC01355 is located at 1p36.12 and contains two exons. Studies on HYPAL are rare, with only Bolun Ai et al. [24] reporting on its role in breast cancer. It indicated LINC01355 acted as a tumor suppressor in breast cancer through FOXO3-mediated CCND1 transcriptional inhibition. Because of the low conservation of lncRNAs and the heterogeneity of the disease, lncRNAs have un-uniformity and even "contradictory" biological functions in different diseases and different cancers. To date, the role of HYPAL in GC has not been reported. Our microarrays showed that downregulating HIF-1α expression caused a significant downregulation of HYPAL. Consequently, we hypothesized that HYPAL expression was regulated by HIF-1α. Subsequent ChIP and luciferase assays confirmed our conjecture, with evidence that HYPAL was regulated by the transcription factor HIF-1α. It confirmed that HYPAL was a veritable hypoxia-associated lncRNA. Analysis of HYPAL expression in GC cells and tissue samples revealed its overexpression in GC, consistent with transcriptome data from the TCGA database. We speculated that HYPAL might be a potential onco-lncRNA in GC based on the microarray and the expression data.
In recent years, considerable attention has been on the molecular regulation of ncRNA, key among them being the functional mechanism of lncRNA. This complex mechanis has been implicated in almost all aspects of gene expression. For example, lncRNA KB-1980E6.3 was shown to act as a molecular scaffold in maintenance of stem cell viability of breast cancer cells by interacting with IGF2BP1 to stabilize c-myc [25]. On the other hand, LncRNA CRNDE was found to reduce chemotherapeutic resistance of GC by regulating the alternative splicing ability of PICALM via SRSF6 [26]. Moreover, numerous pieces of evidences have showed that lncRNAs could act as a molecular sponge to competitively regulate expression of mRNAs[27]. For example, LncRNA AGAP2-AS1 competitively bound to miR-15a-5p to maintain stemness of breast cancer cells and induce trastuzumab resistance [28]. Moreover, LncRNA BCYRN1 suppressed glioma occurrence by competitively regulating CUEDC2 expression[28]. In the present study, bioinformatics analysis of the microarray transcriptome revealed that HYPAL was a potential target molecule for miR-431-5p, while their expression patterns were negatively correlated. In fact, HYPAL promoted GC proliferation, inhibited apoptosis both in vivo and in vitro, and was associated with poor survival of GC patients. However, miR-431-5p antagonized the carcinogenic role of HYPAL. Results from the Luciferase assay showed that miR-431-5p bound to the 3′-UTR of HYAPL to inhibit the expression of HYPAL. Therefore, we further explored the regulatory network for the interaction between HYPAL and miR-431-5p as well as other mRNAs.
The CDK family has been reported to regulate cell cycle transformation. Deregulation of the newly discovered member, CDK14 has been implicated in various cancers. LINC00707 and miR-206 synergistically regulate CDK14 to promote the progression of HCC [29]. Moreover, CDK14 was found to be an oncogene in non-small cell lung cancer cells [30] and glioma [31]. It also participates in mitosis [32] and promotes the progression of various tumors [19, 33] through Wnt/β-catenin signaling pathway. Here, we found that the expression of miR-431-5p and CDK14 was also negatively correlated. In addition, miR-431-5p inhibited the transcription of CDK14 by binding to its 3′-UTR. Low HYPAL expression was accompanied by a significant decrease in the expression level of CDK14. CDK14 promoted the proliferation of GC cells, whereas miR-431-5p inhibited its expression. Further analysis confirmed that HYPAL acted as a ceRNA by sponging miR-431-5p and released the expression of CDK14. In this way, it activated the Wnt/β-catenin signaling pathway to promote the proliferation and inhibit apoptosis of GC cells.
As mentioned above, ncRNAs regulate multiple molecules in cells by directly binding to different regions. Moreover, one gene can be regulated by multiple ncRNAs. It implies that ncRNAs and mRNAs create complex molecular regulatory networks. Our results suggested that HIF-1α activated the HYPAL/miR-431-5p/CDK14 axis in the hypoxic microenvironment of tumors and contributed to the malignant proliferation of GC by activited Wnt/β-catenin pathway. It is the first systematic explanation of the function of HYPAL in GC. And it provides new evidence for changes in GC transcriptomics under hypoxia. However, whether HYPAL regulates the proliferation of GC by interacting with other molecules remain to be determined. Moreover, the pathway through which CDK14 activates Wnt/β-catenin signlaing pathway requires further investigation.
Conclusion
In this study, we originally discovered a novel hypoxia-related lncRNA in GC. It acted as an “accomplice” in the hypoxic microenvironment and assists the proliferation of GC. Mechanistically, HYPAL functions as a competitive sponge for miR-431-5p which activates the CDK14/Wnt/β-catenin pathway thereby induced the GC progression. Our study provides a new approach for identifying potential therapeutic targets in GC.
Availability of data and materials
The data are available from the sources listed in the manuscript—the TCGA data portal. The data used to support the findings of this study are available from the corresponding author upon request.
Abbreviations
- GC:
-
Gastric cancer
- LncRNA:
-
Long non-coding RNAs
- ncRNAs:
-
Non-coding RNAs
- HIF-1α:
-
Hypoxia-inducible factor-1α
- ceRNA:
-
Competing endogenous RNA
- mRNA:
-
Messenger RNA
- HRLs:
-
Hypoxia responsive lncRNAs
- miRNA:
-
MicroRNA
- HYPAL:
-
Hypoxia yield proliferation associated LncRNA
- CDK14:
-
Cyclin-dependent kinase 14
- CDKs:
-
Cyclin-dependent kinases
- ChIP:
-
Chromatin immunoprecipitation
- wt:
-
Wild type
- mut:
-
Mutant type
- sh-HYPAL:
-
Knockdown the expression of HYPAL
- OE-HYPAL:
-
Overexpressed the HYPAL expression
- si-CDK14:
-
Knockdown the expression of CDK14
- OE-CDK14:
-
Overexpressed the CDK14 expression
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: Cancer J Clin. 2020;70:7–30.
Togasaki K, Sugimoto S, Ohta Y, Nanki K, Matano M, Takahashi S, et al. Wnt signaling shapes the histological variation in diffuse gastric cancer. Gastroenterology. 2020;160:823–30.
Zheng ZQ, Chen JT, Zheng MC, Yang LJ, Wang JM, Liu QL, et al. Nestin+/CD31+ cells in the hypoxic perivascular niche regulate glioblastoma chemoresistance by upregulating JAG1 and DLL4. Neuro Oncol. 2020;23:905–19.
Madan E, Peixoto ML, Dimitrion P, Eubank TD, Yekelchyk M, Talukdar S, et al. Cell competition boosts clonal evolution and hypoxic selection in cancer. Trends Cell Biol. 2020;30:967–78.
Zhang J, Guo S, Wu Y, Zheng ZC, Wang Y, Zhao Y. P4HB, a novel hypoxia target gene related to gastric cancer invasion and metastasis. Biomed Res Int. 2019;2019:9749751.
Zhang J, Wu Y, Lin YH, Guo S, Ning PF, Zheng ZC, et al. Prognostic value of hypoxia-inducible factor-1 alpha and prolyl 4-hydroxylase beta polypeptide overexpression in gastric cancer. World J Gastroenterol. 2018;24:2381–91.
Piao HY, Guo S, Wang Y, Zhang J. Exosome-transmitted lncRNA PCGEM1 promotes invasive and metastasis in gastric cancer by maintaining the stability of SNAI1. Clin Transl Oncol. 2020;23:246–56.
Zhang J, Jin HY, Wu Y, Zheng ZC, Guo S, Wang Y, et al. Hypoxia-induced LncRNA PCGEM1 promotes invasion and metastasis of gastric cancer through regulating SNAI1. Clin Transl Oncol. 2019;21:1142–51.
Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J, et al. Translation of non-coding RNAs and cancer. Cancer Lett. 2020;497:89–99.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407.
Pan Y, Fang Y, Xie M, Liu Y, Yu T, Wu X, et al. LINC00675 suppresses cell proliferation and migration via downregulating the H3K4me2 level at the SPRY4 promoter in gastric cancer. Mol Ther Nucleic acids. 2020;22:766–78.
Shuai Y, Ma Z, Liu W, Yu T, Yan C, Jiang H, et al. TEAD4 modulated LncRNA MNX1-AS1 contributes to gastric cancer progression partly through suppressing BTG2 and activating BCL2. Mol Cancer. 2020;19:6.
Chen Z, Chen X, Lu B, Gu Y, Chen Q, Lei T, et al. Up-regulated LINC01234 promotes non-small-cell lung cancer cell metastasis by activating VAV3 and repressing BTG2 expression. J Hematol Oncol. 2020;13:7.
Shih JW, Kung HJ. Long non-coding RNA and tumor hypoxia: new players ushered toward an old arena. J Biomed Sci. 2017;24:53.
Barth DA, Prinz F, Teppan J, Jonas K, Klec C, Pichler M. Long-noncoding RNA (lncRNA) in the regulation of hypoxia-inducible factor (HIF) in cancer. Non-Coding RNA. 2020;6:27.
Hu J, Dang N, Menu E, De Bruyne E, Xu D, Van Camp B, et al. Activation of ATF4 mediates unwanted Mcl-1 accumulation by proteasome inhibition. Blood. 2012;119:826–37.
Yang HJ, Wang L, Wang M, Ma SP, Cheng BF, Li ZC, et al. Serine/threonine-protein kinase PFTK1 modulates oligodendrocyte differentiation via PI3K/AKT pathway. J Mol Neurosci. 2015;55:977–84.
Ou-Yang J, Huang LH, Sun XX. Cyclin-Dependent Kinase 14 promotes cell proliferation, migration and invasion in ovarian cancer by inhibiting Wnt signaling pathway. Gynecol Obstet Invest. 2017;82:230–9.
Li Q, Zhou L, Wang M, Wang N, Li C, Wang J, et al. MicroRNA-613 impedes the proliferation and invasion of glioma cells by targeting cyclin-dependent kinase 14. Biomed Pharmacother. 2018;98:636–42.
Grady WM, Yu M, Markowitz SD. Epigenetic alterations in the gastrointestinal tract: current and emerging use for biomarkers of cancer. Gastroenterology. 2021;160:690–709.
Ramezankhani R, Solhi R, Es HA, Vosough M, Hassan M. Novel molecular targets in gastric adenocarcinoma. Pharmacol Ther. 2020;220:107714.
Tan H, Zhang S, Zhang J, Zhu L, Chen Y, Yang H, et al. Long non-coding RNAs in gastric cancer: new emerging biological functions and therapeutic implications. Theranostics. 2020;10:8880–902.
Li JY, Chen YP, Li YQ, Liu N, Ma J. Chemotherapeutic and targeted agents can modulate the tumor microenvironment and increase the efficacy of immune checkpoint blockades. Mol Cancer. 2021;20:27.
Ai B, Kong X, Wang X, Zhang K, Yang X, Zhai J, et al. LINC01355 suppresses breast cancer growth through FOXO3-mediated transcriptional repression of CCND1. Cell Death Dis. 2019;10:502.
Zhu P, He F, Hou Y, Tu G, Li Q, Jin T, et al. A novel hypoxic long non-coding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021;40(1609):1627.
Zhang F, Wang H, Yu J, Yao X, Yang S, Li W, et al. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer. 2021;20:6.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8.
Han J, Qu H, Han M, Ding Y, Xie M, Hu J, et al. MSC-induced lncRNA AGAP2-AS1 promotes stemness and trastuzumab resistance through regulating CPT1 expression and fatty acid oxidation in breast cancer. Oncogene. 2021;40:833–47.
Tu J, Zhao Z, Xu M, Chen M, Weng Q, Wang J, et al. LINC00707 contributes to hepatocellular carcinoma progression via sponging miR-206 to increase CDK14. J Cell Physiol. 2019;234:10615–24.
Jin B, Jin H, Wu HB, Xu JJ, Li B. Long non-coding RNA SNHG15 promotes CDK14 expression via miR-486 to accelerate non-small cell lung cancer cells progression and metastasis. J Cell Physiol. 2018;233:7164–72.
Zhen Y, Nan Y, Guo S, Zhang L, Li G, Yue S, et al. Knockdown of NEAT1 repressed the malignant progression of glioma through sponging miR-107 and inhibiting CDK14. J Cell Physiol. 2019;234:10671–9.
Kaldis P, Pagano M. Wnt signaling in mitosis. Dev Cell. 2009;17:749–50.
Chen Q, Huang X, Dong X, Wu J, Teng F, Xu H. Long non-coding RNA ERICH3-AS1 is an unfavorable prognostic factor for gastric cancer. PeerJ. 2020;8:e8050.
Acknowledgements
Liaoning S&T Project (2020-ZLLH-33). Jun Zhang especially wishes to thank Taylor Swift, whose songs have given him powerful spiritual support over the past five years.
Author information
Authors and Affiliations
Contributions
HYP performed the majority of experiments and analyzed the data and drafted the manuscript. JZ designed the research. YK and DY conducted the molecular biology assays and assisted in writing the manuscript. YL, YW and XYM collected and analyzed the data.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethics approval and consent to participate
The study was reviewed and approved by the Faculty of Science Ethics Committee at Liaoning Cancer Hospital and Institute (Cancer Hospital of China Medical University, 20181226).
Informed consent
Informed consent was obtained from patients before surgery at the Liaoning Province Cancer Hospital and Institute.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10120_2021_1213_MOESM1_ESM.tif
Supplement Figure 1 GFP fluorescence and RT-PCR showed the efficiency of HIF-1α knockdown compared with that of the scrambled control in GC cells. a. GFP; b. SGC-7901; c. BGC-823 (TIF 8471 KB)
10120_2021_1213_MOESM2_ESM.tif
Supplement Figure 2 Changes in the expression of HYPAL and miR-431-5p. a. The HYPAL shRNAs could inhibited the expression of HYPAL. b. The plasmid could upregulate the expression of HYPAL. c. The mimics and inhibitor of miR-431-5p could change the expression of miR-431-5p correspondingly. d. The mimics and inhibitor of miR-431-5p could effect the GC cells proliferation measured by CCK-8. e. The CDK14 siRNAs could inhibited the expression of CDK14. f. The plasmid could upregulate the expression of CDK14 (TIF 12245 KB)
10120_2021_1213_MOESM3_ESM.tif
Supplement Figure 3 HYPAL reinforced the proliferative capacity of normoxia GC cells. a, b. The mimics of miR-431-5p could reverse the inhibitory effect of OE-HYPAL on normoxia culture GC cell proliferation which were determined CCK-8 assay. c, d. Cell proliferation assessed in HYPAL overexpressed and HYPAL overexpressed + miR-431-5p mimics GC cells by colony formation and EdU assay. e, f. The cell cycle progression of GC cells transfected with OE-NC, OE-HYPAL, OE- HYPAL + miR-431-5p mimics was identified by flow cytometry assay. g, h. Effects of OE-HYPAL, OE-HYPAL + miR-431-5p mimics on GC cell apoptosis (Data are shown as mean ± SD, n = 3. The data statistical significance is assessed by Student's t-test. *P < 0.05) (TIF 22660 KB)
10120_2021_1213_MOESM4_ESM.tif
Supplement Figure 4 CDK14 contributed to the proliferation of normoxia GC cells. a, b. The mimics of miR-431-5p could reverse the promoted effect of OE-CDK14 on normoxia culture GC cell proliferation which were determined CCK-8 assay. c, d. Cell proliferation assessed in CDK14 overexpressed and CDK14 overexpressed + miR-431-5p mimics GC cells by colony formation and EdU assay. e, f. The cell cycle progression of GC cells transfected with OE-NC, OE-CDK14, OE- CDK14 + miR-431-5p mimics was identified by flow cytometry assay. g, h. Effects of OE-CDK14, OE-CDK14 + miR-431-5p mimics on GC cell apoptosis (Data are shown as mean ± SD, n = 3. The data statistical significance is assessed by Student's t-test. *P < 0.05) (TIF 22503 KB)
10120_2021_1213_MOESM5_ESM.tif
Supplement Figure 5 a-f. The statistical results of the change of Wnt/β-catenin pathway protein expression. a. CDK14. b. MMP9. c. c-myc. d. surviving. e. β-catenin. f. pho-β-catenin. g. legend of chart. h. Detected the expression of miR-431-5p in subcutaneous tumor samples (21 days). Increased miR-431-5p expression in miR-431-5p-mimics transfected samples (TIF 11723 KB)
Rights and permissions
About this article
Cite this article
Piao, Hy., Liu, Y., Kang, Y. et al. Hypoxia associated lncRNA HYPAL promotes proliferation of gastric cancer as ceRNA by sponging miR-431-5p to upregulate CDK14. Gastric Cancer 25, 44–63 (2022). https://doi.org/10.1007/s10120-021-01213-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-021-01213-5