Abstract
Objectives
To explore Saccharomyces cerevisiae as an expression platform for dengue oral immune complex vaccine development.
Results
Molecular engineering was applied to create a fusion gene construct (scEDIII-PIGS) consisting of a yeast codon optimized sequence encoding for a synthetic consensus dengue envelope domain III (scEDIII) followed by a modified IgG Fc domain (PIGS). Northern blot showed transcription of the target gene, with a temporal expression pattern similar to those from previous work. Western blot showed assembly of various immune complexes from monomer to hexamer. Partial purification of scEDIII-PIGS was also attempted to demonstrate the feasibility of yeast system for immune complex vaccine development. Approximately 1 mg of scEDIII-PIGS can be produced from 1 l culture.
Conclusion
This work demonstrated for the first time that various immunocomplex structures of our target protein could be efficiently produced in S. cerevisiae for future application in developing oral and injectable vaccines against various pathogens.
Similar content being viewed by others
Introduction
Dengue fever is a disease caused by mosquito-borne dengue virus and becomes a major global health crisis. Dengue viruses consist of four serotypes that are genetically distinct but highly similar (Katzelnick et al. 2015). While infection with one dengue serotype confers life-time protection against that serotype, the most severe forms of dengue fever, dengue hemorrhage fever and dengue shock syndromes, are the result of secondary infection by heterologous serotypes, a process referred to as “antibody-dependent enhancement”, (Balsitis et al. 2010). Although a number of dengue vaccines are being developed, the molecular and immunological reasons behind poor performance are still being studied.
Mucosal vaccines are vaccines that can be delivered nasally, orally, rectally or vaginally. Theoretically, mucosal vaccines are cheaper, safer, and more appropriate for mass distribution than parenteral vaccines (Lycke 2012; Zeng 2016; Miquel-Clopés et al. 2019).
Among various delivery systems for mucosal vaccines, Saccharomyces cerevisiae represents a very attractive choice. Yeast is a GRAS organism and long used as a food supplement. Moreover, S. cerevisiae is a cost-effective eukaryotic heterologous expression host. Finally, yeast cells are sufficiently immunogenic in human as a result of carbohydrate molecules on their cell wall that act as Pathogen Associated Molecular Patterns (PAMPs) (Kumar and Kumar 2019).
Immunocomplexes (ICs) are formed when antibodies like IgG or IgM bind to their cognate antigens. ICs may work as vaccines, show superior immunogenicity compared with alum-precipitated antigens, and also induce stronger cellular and humoral immune responses (Hioe et al. 2009; Kumar et al. 2013). Recombinant DNA technology allows for convenient production of recombinant ICs in which antigen fused Fc fragments are capable of forming more complex structures. These structures are collectively referred to as PIGS (Polymeric IgG Scaffolds).
Although several expression hosts, including plant and CHO cells, have been used to express PIGS-based antigens (Kim et al. 2017, 2018), no study has used yeast as an expression host for PIGS. In this study, we explored the possibility of using S. cerevisiae to produce an immunocomplex (scEDIII-PIGS) comprised of a synthetic consensus dengue envelope domain III (scEDIII) conjugated with a murine Fc fragment from IgG2 (PIGS). The scope of this study is limited to the expression and purification of the scEDIII-PIGS for subsequent use in the development of dengue vaccine.
Materials and methods
Strains and culture conditions
All plasmids used in this work were maintained in Escherichia coli Top10 strain. BL21 (de3) RIPL was used for the production of synthetic consensus Dengue envelope domain III antigen (scEDIII) in E. coli. The E. coli strains were maintained in Luria Bertani broth supplemented with appropriate antibiotics. The Saccharomyces cerevisiae 2805 strain (MATα pep4::HIS3 prb 1-δ Can1 GAL2 his3 ura3-52) used previously by our group in other yeast-based oral vaccine development work (Kim et al. 2011; Bal et al. 2018a, b) was used as the host for scEDIII-Fc complexes (scEDIII-PIGS). The recipient strain was maintained in YEPD liquid and agar media. The transformants were maintained in uracil dropout media (Ura−), which was comprised of yeast nitrogen base with ammonium sulfate (0.56% w/v), KCl (0.76% w/v), glucose (2% w/v), and yeast synthetic dropout (0.14% w/v), and supplemented with 20 mg of tryptophan, histidine, leucine, and adenine hemisulfate.
Escherichia coli and Saccharomyces cerevisiae chemical transformations were employed as previously described (Nguyen et al. 2013; Bal et al. 2018a). For scEDIII-PIGS production, a single colony of the transformant was inoculated into 5 ml of Ura− liquid medium for 48 h in a shaking incubator at 200 rpm and 30 °C. 250 µl of this seed culture was transferred into 5 ml of YEPD medium and cultured for 16 h in identical conditions. This culture was subsequently inoculated into a baffled Erlenmeyer 300 ml flask containing 40 ml of YEPD medium and further cultured until cells were harvested for expression analysis.
Construction of the expression vector
The expression construct encoding for the scEDIII-PIGS was created by overlap extension PCR between the gene encoding yeast codon-optimized scEDIII (GenBank ID: DI43185731, Nguyen et al. 2013) and the gene encoding murine-modified IgG2a-Fc fragment (Kim et al. 2017). The fusion construct was cloned into a pGEM-T Easy Vector system and its sequence confirmed. This fusion construct was subsequently cloned into the yeast episomal vector pYEGPD-TER (Lim et al. 2003) using BamHI and SalI restriction enzymes. The resulting recombinant vector (pYEGPD-scEDIII-PIGS) was sequenced with a primer pair flanking the expression construct and the result was compared with the reference sequence to ensure the accuracy of the ORF (Supplementary Fig. 1 and Supplementary Table 1).
Colony PCR and back transformation
Colony PCR was used to confirm the presence of the pYEGPD-scEDIII-PIGS episomal plasmid. Escherichia coli back transformation was employed to ascertain the integrity of the expression cassette after yeast transformation as described previously (Nguyen et al. 2013).
Expression analysis
The expression of scEDIII-PIGS was examined at the transcription and translation level. Northern blot analysis was conducted to detect the accumulation of the scEDIII-PIGS transcript. For the Northern blot analysis, 20 randomly selected transformants were cultured for three days and their total RNA was analyzed for the presence of the scEDIII-PIGS mRNA to determine the transformants with the highest transcription levels. The two transformants displaying the strongest hybridizing bands were selected and subsequently analyzed for their temporal expression pattern at the transcription and translation levels (1-, 3-, and 5-days post-inoculation).
Northern blot analysis
Total yeast RNA preparation was carried out using the procedure described by Lim et al. (2003). RNA concentrations were determined using 96 microplates in the Multiskan GO UV spectrophotometer (Thermo Fisher Scientific Inc.) and approximately 30 µg of total RNA from each yeast transformant was analyzed on 1.2% (w/v) denatured formaldehyde-agarose gel. RNAs were blotted onto an Amersham Hybond™ nylon membrane (GE Healthcare) according to the manufacturer’s instructions. Hybridization was carried out in modified Church buffer (Nguyen et al. 2013) with a scEDIII-PIGS PCR product as a detecting probe.
Protein preparation, SDS-PAGE and Western blot analysis
Approximately 500 µl of wet cell mass was mixed with 400 µl of 0.5-mm diameter acid washed glass bead (BioSpec Products Inc.) and 600 µl of breaking buffer (200 mM Tris–HCl pH 8.0, 150 mM ammonium sulfate, 1 mM EDTA and 10% v/v glycerol). The mixture was homogenized using FastPrep24™ 5G bead beater (MP Biomedicals) by beating for 30 s 8 times with a 1-min cooling interval between beatings. The lysate was first centrifuged at 7000 rpm for 20 min at 4 °C and the supernatant was collected and further clarified by 20-min centrifugation at 13,000 rpm. After concentrations were determined by Bradford assay, the protein samples were stored at − 20 °C for subsequent analysis.
For SDS-PAGE and Western blot analysis approximately 100 µg or 200 µg of total protein from each putative transformant was analyzed on a discontinuous SDS-PAGE gel consisting of 5% (v/v) stacking gel and 10% or 12% (v/v) separating gel. For samples in a denatured condition, samples were boiled for 10 min prior to the loading on the gel. For samples in a non-denatured condition, samples were loaded on the gel after being mixed with 6 × loading buffer without dithiothreitol (non-reducing 6 × loading buffer). The gels were subsequently blotted onto a 0.4 µm Nylon membrane and the target protein was detected by anti-dengue antibody (LSBio).
Purification of the target proteins
Functional monomeric and polymeric scEDIII-PIGS were purified at a small scale using Pierce Protein A agarose (Thermo Fisher Scientific Inc.). Eluent fractions were collected in 1 ml each and subsequently concentrated with either 5 × (v/v) cold acetone or Amicon Ultra-15 centrifugal filter (Merck Millipore). Alternatively, the proteins were purified with Bioneer’s Maglisto magnetic protein A kit. The eluents were analyzed on 8% (v/v) SDS-PAGE gel in a non-denatured condition (without boiling in non-reducing 6 × loading buffer) and the gel was blotted on a Nylon membrane. Purified and active scEDIII-PIGS were detected as described above.
Results
Cloning and characterization of the expression cassette
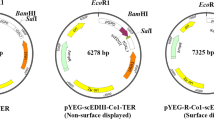
In this study, the episomal vector yEPGPD-TER, which has already been used to express various antigens in yeast (Kim et al. 2011; Nguyen et al. 2013, 2015; Bal et al. 2018a, b), was employed to express the target gene. This vector is a high copy number vector containing a 2 µ yeast replication origin sequence that allows the vector to replicate independently from chromosomal DNA. The scEDIII-PIGS construct consists of a yeast codon-optimized gene that encodes a proof of concept tetravalent dengue antigen (Nguyen et al. 2013) fused with a sequence that encodes a mouse IgG2 Fc fragment. The Fc fragment contains extra amino acids at the N terminus, crossing the hinge region and the final ten amino acids of the CH1 domain, as well as an IgM µ tail piece (µtp) at the C-terminus. Furthermore, a Pro476 to Thr476 substitution (Pro476Thr) was performed in order to make the final three or four amino acids at the C-terminus of the IgG2 Fc fragment identical to those of the IgM Fc fragment (Kim et al. 2017). The scEDIII-PIGS construct is under the control of the strong constitutive glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter and the galactose-1-phosphate uridylyltransferase (GAL7) terminator (Fig. 1).
Expression and assembly of the target gene product
Transformed yeast cells were selected on the Ura− medium. The transformation efficiency was extremely high, with 10 µl of competent cells yielding more than 100 putative transformants per plate. When the putative transformants were analyzed by colony PCR, all transformants showed an expected band at approximately 1 kb with the exception of the mock transformant (data not shown). Escherichia coli back transformation and subsequent sequence analysis of the pYEGPD-scEDIII-PIGS showed that the integrity of the expression cassette was preserved during the chemical transformation (data not shown). Northern blot analysis (Fig. 2) of 20 randomly selected putative transformants revealed successful transcription of the target gene in all examined transformants. Based on the Northern blot analysis, two strains with the strongest transcription levels were selected for their temporal expression pattern. The results showed that the scEDIII-PIGS transcription level was highest at day 1 and then decreased rapidly at day 3 post-inoculation (Fig. 3). This is consistent with our previous results (Nguyen et al. 2013; Bal et al. 2018a) and may be explained by the fact that in nonselective media, episomal plasmids will gradually dilute out.
Northern blot analysis of scEDIII-PIGS transcript. 20 randomly selected transformants were analyzed. Above: Northern blot signals showing the transcription of scEDIII-PIGS expression cassette. Bellow: rRNA electrophoresis figure was shown to indicate equivalent amount of total RNA for each transformant was compared. Each lane contained 30 µg of total RNA. From this result, two strongly expressed strains (transformants #13 and #17) were selected for further analysis
Temporal expression pattern of scEDIII-PIGS transcripts from two selected transformants #13 and #17. (Lanes 1, 2, and 3): total RNA from strain #13 at 1-, 3-, and 5-days post-inoculation; (Lanes 4, 5, and 6): total RNA from strain #17 at 1-, 3-, and 5-days post-inoculation. Each yeast sample analyzed in this experiment contained 30 µg of total RNA. rRNA electrophoresis bands are shown to indicate equal amount of total RNA
Western blot analysis of the cell free extracts from the selected two strains with the strongest transcription at different time points (1-, 3-, and 5-day post-inoculation) displayed the distinctive antibody-reacting band at the expected size of 40 kDa under the denatured (sample-boiling) conditions, corresponding to the single chain of scEDIII-PIGS (Fig. 4a, b). Without question, the protein product of the target gene was successfully expressed. Although analysis of deduced amino acid sequences of scEDIII-PIGS did reveal the presence of two canonical N-glycosylation sites (Asn-X-Ser/Thr) at amino acid residues of 196–198 and 351–353, the lack of a significant difference in the size of the single chain of scEDIII-PIGS from the estimated size on the gel, in conjunction with the absence of microheterogeneity of expressed scEDIII-PIGS, suggested that glycosylation may not be strong in scEDIII-PIGS. Consistent with the Northern blot analysis, protein production (represented by the intensity of antibody-specific bands at 40 kDa under denatured condition) was strongest at day 1 and weaker by day 3 and day 5 (Supplementary Table 2). Based on the relative intensity of the antibody binding band in the Western blot, we estimated that 1 l of yeast culture yields approximately 1 mg of scEDIII-PIGS.
Temporal expression analysis by Western blot of two selected strains (transformants #13 and #17) under denatured (boiling) condition (a and b) and nondenatured (non-boiling) condition (c and d). SDS-PAGE images (b and d) are shown to indicate equal amount of protein for each sample. M: Pageruler prestained protein ladder (Thermo Fisher Scientific Inc.). a and b lane 1: 250 ng of purified E. coli expressed scEDIII; lanes 2, 3, and 4: total soluble protein from transformant #13 at 1-, 3-, and 5-days post-inoculation, respectively; lanes 5, 6, and 7: total soluble protein from transformant #17 at 1-, 3-, and 5-days post-inoculation, respectively; lane 8: total soluble protein from mock transformant. c and d: lane 1: total soluble protein from mock transformant; lanes 2, 3, and 4: total soluble protein from transformant #13 at 1-, 3-, and 5-days post-inoculation; lanes 5, 6 and 7: total soluble protein from transformant #17 at 1-, 3-, and 5-days post-inoculation; lane 8: 250 ng of purified E. coli expressed scEDIII. All yeast samples contain equal amount (100 µg in a and b and 200 µg in c and d) of total soluble protein. Arrows point to the positions of single chain scEDIII-PIGS (lower) while brackets (upper) indicate monomeric to polymeric (putatively pentameric/hexameric) forms on SDS-PAGE gels
More importantly, in non-denatured condition, several additional bands that correspond to single chain scEDIII-PIGS (~ 40 kDa), monomeric scEDIII-PIGS (~ 80 kDa), and polymeric scEDIII-PIGS (~ 160 to 480 kDa) were observed (Fig. 4c, d). The band intensity of these polymeric scEDIII-PIGS forms was more distinct at day 1 than at days 3 and day 5, which also suggests increased expression and assembly at the early expression stage. Antibody-specific binding, and the appearance of single and multiple band patterns depending on the denatured- and non-denatured condition of sample proteins are distinct characteristics of PIGS-based expression, and have been previously described (Kim et al. 2017, 2018). This further confirms that yeast-expressed scEDIII-PIGS are expressed well and, moreover, are able to be assembled into polymeric structures when in a non-denatured condition.
Purification of scEDIII-PIGS and evaluation of their potential for vaccine development
scEDIII-PIGS purification relies on the premise that scEDIII-PIGS behave like immunoglobulins due to their common Fc fragment. Indeed, we were able to partially purify scEDIII-PIGS to using either protein A conjugated magnetic beads or protein A conjugated agarose gravity flow columns. SDS-PAGE and Western blot analysis showed a distinct band at approximately 100 kDa, which probably correspond to the monomeric scEDIII-PIGS, and an intense smearing above that band but not below it, which corresponds to dimeric, pentameric, and hexameric forms (Fig. 5). This smearing pattern of high molecular weight forms of scEDIII-PIGS agreed with previous works expressing scEDIII-PIGS in tobacco and animal cells (Kim et al. 2017, 2018).
Western blot analysis of scEDIII-PIGS purification. a SDS-PAGE analysis of samples under non-denatured conditions; b Western blot analysis from the corresponding SDS-PAGE gel. Lane 1: protein ladder (Thermo Fisher Scientific Inc.); lane 2: Elution fraction 3; lane 3: Elution fraction 2; lane 4: elution fraction 1; lane 5: flowthrough; and lane 6: total protein. Brackets (upper) and an arrow (lower) indicate monomeric to polymeric (putatively pentameric/hexameric) forms and single chain scEDIII-PIGS, respectively, on SDS-PAGE gels
Discussion
Monoclonal antibody (mAb) production has revolutionized biotechnology, with mAbs currently accounting for 40% of the global biopharmaceutical market (https://www.datamintelligence.com/research-report/biopharmaceuticals-market). While a great deal of research has focused on the antigen binding portion of antibodies, attention is increasingly being paid to the possibility of exploiting the biological functions of the Fc fragment (DiLillo et al. 2015; Blundell et al. 2017; Mackness et al. 2019; Kariolis et al. 2020). New classes of biopharmaceuticals include therapeutic Fc-fusion proteins, which improve half-life and efficiency. To date, thirteen such biopharmaceuticals have been approved in Europe and the United States (Duivelshof et al. 2020). Interest in the Fc fragment has also been focused on the potential use of the immunocomplex to vaccinate against various infectious diseases and contribute the fight against cancers (Hioe et al. 2009; Kumar et al. 2013; DiLillo et al. 2015; Kim et al. 2015; Kim et al. 2017).
Mammalian bodies contain five classes of immunoglobulin; IgM, IgG, IgA, IgE and IgD. The Fc fragment associated with each isotype distinguishes each from the others and confers its distinct biological functions. IgMs are the first isotype that are secreted by B cells (Kubagawa et al. 2019). They have low antigen affinity but high avidity due to their polymeric nature. IgMs can assemble into pentamers in the presence of J chain or hexamer in the absence of J chain (Brewer and Corley 1997). In-depth study of the principles of IgM hexamer formation showed that CH4 and the µtp are sufficient preconditions for its hexameric formation (Müller et al. 2013). Interest in IgMs grew once it was proposed that it might be possible to engineer Fc from IgG, and that this might result in a hybrid IgM-IgG Fc that allowed for multimeric assembly and possessed the desirable properties of both IgM and IgG (Smith and Morrison 1994). Previous studies created hexameric IgG by grafting a modified IgG C-terminal tail piece to the IgG CH3 domain. Ultimately, this process resulted in the successful production of multimeric sCEDIII-PIGS in Nicotiana benthamiana and CHO cells (Kim et al. 2017).
Vaccines are hailed as one of the most important discoveries of the previous century, and have saved millions of lives. Unfortunately, there remain many diseases where vaccines are either unavailable or ineffective, including HIV, malaria, and cancer. Dengue is currently the most serious mosquito borne infection (Diamond and Pierson 2015), and while some dengue vaccines have reached the clinical trial stage and even received regulatory approval, no vaccine has gained universal acceptance (Idris et al. 2020). With the ongoing SARS-CoV-2 pandemic taking up so many public health resources, the imperative of improving the vaccine development process to prevent other infectious diseases is very clear. An immunocomplex approach such as scEDIII-PIGS has significant potential as a novel vaccine development approach for a number of diseases, but particularly for the development of an oral Dengue vaccine.
Although expression using S. cerevisiae has the same intrinsic limits associated with all heterologous proteins, i.e., questioning the assembly authenticity of the expressed protein and posttranslational modification, it offers significant advantages to vaccine development over other expression systems, as yeast cells are suitable to be taken up by antigen-presenting cells (APCs) (Bernstein et al. 2008) and are potent transporters into dendritic cells, triggering antigen-specific CD4 + and CD8 + immune responses in vivo (Howland et al. 2008; Liu et al. 2016). In addition, S. cerevisiae tends to trigger potent innate as well as adaptive T cell immune responses (Bucarey et al. 2009; Bal et al. 2018a). In our previous papers, we used a yeast-expressed synthetic consensus sequence of EDIII (scEDIII) as a proof-of-concept Dengue tetravalent antigen to test various strategies for developing tetravalent Dengue vaccines. We showed that purified scEDIII from recombinant yeast induced a balanced immune response in mice and that when it was conjugated with gut mucosal layer binding ligands such as Co1 or LTB, it had immense potential as a recombinant tetravalent Dengue oral vaccine (Nguyen et al. 2013; Bal et al. 2018a, b). Building on our success of strong immunogenicity and neutralizing activity we previously achieved using yeast-expressed scEDIII, in this work we attempted to produce an immune-complex-like structure (scEDIII-PIGS) using S. cerevisiae. The simplicity of this system, which consists of an episomal vector (yEPGDP-TER) transformed into an auxotrophic mutant yeast strain (2085, ura3−) using LiAc method, means that various strategies for vaccine development can be attempted at very low cost and in a very time-efficient manner. Although it is clear from the Western results that immune complexes have formed successfully, the expression level can still be improved further using different combinations of promoters and terminators since we can see from the Northern blot results that the transcription level of the target gene dropped markedly over time. Expression level is a crucial factor since this synthetic complex is intended to work as a Dengue oral vaccine candidate. The appearance of the monomeric product at approximately 100 kDa may reflect the atypical movement of the protein in SDS-PAGE under non-reducing conditions instead of changes in molecular weight since the single chain scEDIII-PIGS moved with expected molecular weight. Indeed, under non-boiling but reducing conditions, we observed distinct bands at expected sizes (data not shown).
Our expression level of scEDIII-PIGS in yeast system appears to be comparable to the same protein expressed in N. benthamiana and CHO cells by Kim et al. (2017). While the same protein construct expressed in plants and CHO cells have been previously shown to work extremely well as a parenteral vaccine (Kim et al. 2017, 2018), it still required alum adjuvant. Apparently, injection is not desirable for dengue vaccines as resource limited countries simply lack the capacity to successfully implement an injection-based mass dengue vaccination program. An intrinsically adjuvanted, cheap, and convenient system such as S cerevisiae, however, which produces immune-complex at a high expression level and offers a various customizable genetic toolkit that permits further engineering, can form the basis of an effective oral vaccine.
Conclusion
This study shows for the first time that various immunocomplex structures of our target protein, ranging from monomer to hexamer, can be efficiently produced in S. cerevisiae. We cloned a fusion gene construct encoding for murine scEDIII-PIGS using a high-copy yeast episomal expression vector, pYEGPD-TER, which we used to transform the S. cerevisiae 2805 strain. Successful expression was observed via Northern and Western blot analysis. Temporal expression analysis showed that scEDIII-PIGS expression peaked on day one and decreased over time, possibly due to the loss of the episomal vector under a non-selective condition. Notably, the Western results revealed the presence of polymeric scEDIII-PIGS, which can be purified using protein A affinity chromatography. The final yield of scEDIII-PIGS is estimated to be approximately 1 mg per liter of culture. scEDIII-PIGS, which are potent immune complex dengue vaccines, are produced in S. cerevisiae at a level that would potentially warrant mass adoption of this method and allow for the widespread distribution of this novel dengue vaccine. The resulting vaccine may be delivered orally as crude cell extract or parenterally as purified immune complexes.
References
Bal J, Jung HY, Nguyen LN et al (2018a) Evaluation of cell-surface displayed synthetic consensus dengue EDIII cells as a potent oral vaccine candidate. Microb Cell Fact 17:146. https://doi.org/10.1186/s12934-018-0994-8
Bal J, Nguyen LN, Park JC et al (2018b) Comparative immunogenicity of preparations of yeast-derived dengue oral vaccine candidate. Microb Cell Fact 17:24. https://doi.org/10.1186/s12934-018-0876-0
Balsitis SJ, Williams KL, Lachica R et al (2010) Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog 6:e1000790. https://doi.org/10.1371/journal.ppat.1000790
Bernstein MB, Chakraborty M, Wansley EK et al (2008) Recombinant Saccharomyces cerevisiae (yeast-CEA) as a potent activator of murine dendritic cells. Vaccine 26:509–521. https://doi.org/10.1016/j.vaccine.2007.11.033
Blundell PA, Le NPL, Allen J et al (2017) Engineering the fragment crystallizable (Fc) region of human IgG1 multimers and monomers to fine tune interaction with sialic acid-dependent receptors. J Biol Chem 292:12994–13007. https://doi.org/10.1074/jbc.M117.795047
Brewer JW, Corley RB (1997) Late events in assembly determine the polymeric structure and biological activity of secretory IgM. Mol Immunol 34:323–331. https://doi.org/10.1016/s0161-5890(97)00029-1
Bucarey SA, Noriega J, Reyes P et al (2009) The optimized capsid gene of porcine circovirus type 2 expressed in yeast forms virus-like particles and elicits antibody responses in mice fed with recombinant yeast extracts. Vaccine 27:5781–5790. https://doi.org/10.1016/j.vaccine.2009.07.061
Diamond MS, Pierson TC (2015) Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell 162:488–492. https://doi.org/10.1016/j.cell.2015.07.005
DiLillo DJ, Ravetch JV (2015) Differential Fc-receptor engagement drives an anti-tumor vaccinal effect. Cell 161:1035–1045. https://doi.org/10.1016/j.cell.2015.04.016
Duivelshof BL, Murisier A, Camperi J et al (2020) Therapeutic Fc-fusion proteins: current analytical strategies. J Sep Sci. https://doi.org/10.1002/jssc.202000765
Hioe CE, Visciano ML, Kumar R et al (2009) The use of immune complex vaccines to enhance antibody responses against neutralizing epitopes on HIV-1 envelope gp120. Vaccine 28:352–360. https://doi.org/10.1016/j.vaccine.2009.10.040
Howland SW, Tsuji T, Gnjatic S et al (2008) Inducing efficient cross-priming using antigen-coated yeast particles. J Immunother 31:607–619. https://doi.org/10.1097/CJI.0b013e318181c87f
Idris F, Ting DHR, Alonso S (2020) An update on dengue vaccine development, challenges, and future perspectives. Expert Opin Durg Discov 25:1–12. https://doi.org/10.1080/17460441.2020.1811675
Katzelnick LC, Fonville JM, Gromowski GD et al (2015) Dengue viruses cluster antigenically but not as discrete serotypes. Science 349:1338–1343. https://doi.org/10.1126/science.aac5017
Kariolis MS, Wells RC, Getz JA et al (2020) Brain delivery of therapeutic proteins using an Fc fragment blood-brain barrier transport vehicle in mice and monkeys. Sci Transl Med 12:eaay1359. https://doi.org/10.1126/scitranslmed.aay1359
Kim JM, Park SM, Kim JA et al (2011) Functional pentameric formation via coexpression of the Escherichia coli heat-labile enterotoxin B subunit and its fusion protein subunit with a neutralizing epitope of ApxIIA exotoxin improves the mucosal immunogenicity and protection against challenge by Actinobacillus pleuropneumoniae. Clin Vaccine Immunol 18:2168–2177. https://doi.org/10.1128/CVI.05230-11
Kim M-Y, Reljic R, Kilbourne J et al (2015) Novel vaccination approach for dengue infection based on recombinant immune complex universal platform. Vaccine 33:1830–1838. https://doi.org/10.1016/j.vaccine.2015.02.036
Kim M-Y, Van Dolleweerd C, Copland A et al (2017) Molecular engineering and plant expression of an immunoglobulin heavy chain scaffold for delivery of a dengue vaccine candidate. Plant Biotechnol J 15:1590–1601. https://doi.org/10.1111/pbi.12741
Kim MY, Copland A, Nayak K et al (2018) Plant-expressed Fc-fusion protein tetravalent dengue vaccine with inherent adjuvant properties. Plant Biotechnol J 16:1283–1294. https://doi.org/10.1111/pbi.12869
Kubagawa H, Honjo K, Ohkura N et al (2019) Functional roles of the IgM Fc receptor in the immune system. Front Immunol 10:945. https://doi.org/10.3389/fimmu.2019.00945
Kumar R, Kumar P (2019) Yeast-based vaccines: New perspective in vaccine development and application. FEMS Yeast Research 19:foz007. https://doi.org/10.1093/femsyr/foz007
Kumar R, Tuen M, Liu J et al (2013) Elicitation of broadly reactive antibodies against glycan-modulated neutralizing V3 epitopes of HIV-1 by immune complex vaccines. Vaccine 31:5413–5421. https://doi.org/10.1016/j.vaccine.2013.09.010
Lim YY, Park SM, Jang YS et al (2003) Production of a functional mouse interferon γ from recombinant Saccharomyces cerevisiae. J Microbiol Biotechn 13:537–543
Liu Z, Zhou G, Ren C et al (2016) Oral administration of myostatin-specific recombinant Saccharomyces cerevisiae vaccine in rabbit. Vaccine 34:2378–2382. https://doi.org/10.1016/j.vaccine.2016.03.036
Lycke N (2012) Recent progress in mucosal vaccine development: potential and limitations. Nat Rev Immunol 12:592–605. https://doi.org/10.1038/nri3251
Mackness BC, Jaworski JA, Boudanova E et al (2019) Antibody Fc engineering for enhanced neonatal Fc receptor binding and prolonged circulation half-life. mAbs 11:1276–1288. https://doi.org/10.1080/19420862.2019.1633883
Miquel-Clopés A, Bentley EG, Stewart JP, Carding SR (2019) Mucosal vaccines and technology. Clin Exp Immunol 192:205–214. https://doi.org/10.1111/cei.13285
Müller R, Grawert MA, Kern T et al (2013) High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation. Proc Natl Acad Sci USA 110:10183–10188. https://doi.org/10.1073/pnas.1300547110
Nguyen NL, Kim JM, Park JA et al (2013) Expression and purification of an immunogenic dengue virus epitope using a synthetic consensus sequence of envelope domain III and Saccharomyces cerevisiae. Protein Expr Purif 88:235–242. https://doi.org/10.1016/j.pep.2013.01.009
Nguyen NL, So KK, Kim JM et al (2015) Expression and characterization of an M cell-specific ligand-fused dengue virus tetravalent epitope using Saccharomyces cerevisiae. J Biosci Bioeng 119:19–27. https://doi.org/10.1016/j.jbiosc.2014.06.005
Smith RI, Morrison SL (1994) Recombinant polymeric IgG: an approach to engineering more potent antibodies. Nat Biotechnol 12:683–688. https://doi.org/10.1038/nbt0794-683
Zeng L (2016) Mucosal adjuvants: opportunities and challenges. Hum Vaccin Immunother 12:2456–2458. https://doi.org/10.1080/21645515.2016.1181236
Funding
This work was supported in part by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1A6A1A03015876). We are thankful to the Institute of Molecular Biology and Genetics at Jeonbuk National University for kindly providing the facilities for this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
So, KK., Chun, J., Luong, N.N. et al. Expression of an immunocomplex consisting of Fc fragment fused with a consensus dengue envelope domain III in Saccharomyces cerevisiae. Biotechnol Lett 43, 1895–1904 (2021). https://doi.org/10.1007/s10529-021-03161-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-021-03161-7