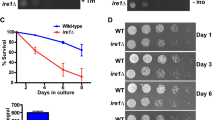
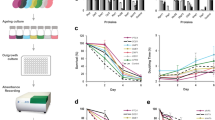
Abstract
Calorie restriction (CR) is the most robust longevity intervention, extending lifespan from yeast to mammals. Numerous conserved pathways regulating aging and mediating CR have been identified; however, the overall proteomic changes during these conditions remain largely unexplored. We compared proteomes between young and replicatively aged yeast cells under normal and CR conditions using the Stable-Isotope Labeling by Amino acids in Cell culture (SILAC) quantitative proteomics and discovered distinct signatures in the aging proteome. We found remarkable proteomic similarities between aged and CR cells, including induction of stress response pathways, providing evidence that CR pathways are engaged in aged cells. These observations also uncovered aberrant changes in mitochondria membrane proteins as well as a proteolytic cellular state in old cells. These proteomics analyses help identify potential genes and pathways that have causal effects on longevity.





Similar content being viewed by others
Availability of data and material
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018917. The RNA-seq data have been deposited to SRA under accession number PRJNA715646.
All experimental materials generated by this study are available upon request.
Code availability
Not applicable.
References
Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459(7248):802–7. https://doi.org/10.1038/nature08085.
Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449(7164):928–32. https://doi.org/10.1038/nature06160.
Rappsilber J, Mann M. Analysis of the topology of protein complexes using cross-linking and mass spectrometry. CSH Protoc. 2007;2007:pdb prot4594. doi:https://doi.org/10.1101/pdb.prot4594.
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26(12):1367–72. doi:https://doi.org/10.1038/nbt.1511.
Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–805. https://doi.org/10.1021/pr101065j.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. https://doi.org/10.1038/nprot.2008.211.
Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R et al. (2009) Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics, Chapter 13:Unit 13 1. doi:https://doi.org/10.1002/0471250953.bi1311s27.
Oughtred R, Stark C, Breitkreutz B-J, Rust J, Boucher L, Chang C, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–41. https://doi.org/10.1093/nar/gky1079.
Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296(5569):910–3.
Jo MC, Liu W, Gu L, Dang W, Qin L. High-throughput analysis of yeast replicative aging using a microfluidic system. Proc Natl Acad Sci U S A. 2015;112(30):9364–9. https://doi.org/10.1073/pnas.1510328112.
Guarente L. Mitochondria–a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132(2):171–6. https://doi.org/10.1016/j.cell.2008.01.007.
Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett. 2011;585(13):2041–8. https://doi.org/10.1016/j.febslet.2010.11.016.
Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464(7288):520–8. https://doi.org/10.1038/nature08982.
Kourtis N, Tavernarakis N. Cellular stress response pathways and ageing: intricate molecular relationships. EMBO J. 2011;30(13):2520–31. https://doi.org/10.1038/emboj.2011.162.
Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mech Ageing Dev. 2005;126(3):365–79. https://doi.org/10.1016/j.mad.2004.08.012.
Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res Rev. 2011;10(2):205–15. https://doi.org/10.1016/j.arr.2010.02.001.
Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333(6046):1109–12. https://doi.org/10.1126/science.1201940.
Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–29. https://doi.org/10.1016/j.cell.2005.11.044.
Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007;35(22):7466–74. https://doi.org/10.1093/nar/gkm756.
Lin SS, Manchester JK, Gordon JI. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J Biol Chem. 2001;276(38):36000–7. https://doi.org/10.1074/jbc.M103509200.
Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12(18):1566–73.
Lesur I, Campbell JL. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Mol Biol Cell. 2004;15(3):1297–312. https://doi.org/10.1091/mbc.e03-10-0742.
Laun P, Ramachandran L, Jarolim S, Herker E, Liang P, Wang J, et al. A comparison of the aging and apoptotic transcriptome of Saccharomyces cerevisiae. FEMS Yeast Res. 2005;5(12):1261–72. https://doi.org/10.1016/j.femsyr.2005.07.006.
Zhan M, Yamaza H, Sun Y, Sinclair J, Li H, Zou S. Temporal and spatial transcriptional profiles of aging in Drosophila melanogaster. Genome Res. 2007;17(8):1236–43. https://doi.org/10.1101/gr.6216607.
de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455(7217):1251–4. https://doi.org/10.1038/nature07341.
Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, et al. Elevated histone expression promotes life span extension. Mol Cell. 2010;39(5):724–35. https://doi.org/10.1016/j.molcel.2010.08.015.
Xie F, Liu T, Qian WJ, Petyuk VA, Smith RD. Liquid chromatography-mass spectrometry-based quantitative proteomics. J Biol Chem. 2011;286(29):25443–9. https://doi.org/10.1074/jbc.R110.199703.
Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–99. https://doi.org/10.1146/annurev-biochem-061308-093216.
Walther DM, Mann M (2011) Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol Cell Proteomics 10(2):M110 004523. doi:https://doi.org/10.1074/mcp.M110.004523.
Jazwinski SM. Yeast longevity and aging–the mitochondrial connection. Mech Ageing Dev. 2005;126(2):243–8. https://doi.org/10.1016/j.mad.2004.08.016.
Thorpe PH, Bruno J, Rothstein R. Modeling stem cell asymmetry in yeast. Cold Spring Harb Symp Quant Biol. 2008;73:81–8. https://doi.org/10.1101/sqb.2008.73.010.
Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–54. https://doi.org/10.1146/annurev.biochem.77.061206.171059.
Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310(5751):1193–6. https://doi.org/10.1126/science.1115535.
Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133(2):292–302. https://doi.org/10.1016/j.cell.2008.02.037.
Vaquero A, Reinberg D. Calorie restriction and the exercise of chromatin. Genes Dev. 2009;23(16):1849–69. https://doi.org/10.1101/gad.1807009.
Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289(5487):2126–8.
Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18(1):12–6. https://doi.org/10.1101/gad.1164804.
Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418(6895):344–8. https://doi.org/10.1038/nature00829.
Molin M, Yang J, Hanzen S, Toledano MB, Labarre J, Nystrom T. Life span extension and H(2)O(2) resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol Cell. 2011;43(5):823–33. https://doi.org/10.1016/j.molcel.2011.07.027.
Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radic Biol Med. 2011;51(2):327–36. https://doi.org/10.1016/j.freeradbiomed.2011.05.010.
Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–80. https://doi.org/10.1016/j.cell.2010.08.020.
Rattan SI. Synthesis, modification and turnover of proteins during aging. Adv Exp Med Biol. 2010;694:1–13.
Powell CD, Quain DE, Smart KA. The impact of brewing yeast cell age on fermentation performance, attenuation and flocculation. FEMS Yeast Res. 2003;3(2):149–57. https://doi.org/10.1016/S1567-1356(03)00002-3.
Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21(19):2410–21. https://doi.org/10.1101/gad.439307.
Orlandi I, Bettiga M, Alberghina L, Nystrom T, Vai M. Sir2-dependent asymmetric segregation of damaged proteins in ubp10 null mutants is independent of genomic silencing. Biochim Biophys Acta. 2010;1803(5):630–8. https://doi.org/10.1016/j.bbamcr.2010.02.009.
Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464(7288):513–9. https://doi.org/10.1038/nature08981.
Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009;37(12):3969–80. https://doi.org/10.1093/nar/gkp270.
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. https://doi.org/10.1016/j.cell.2013.05.039.
Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21(12):1406–15. https://doi.org/10.1038/nm.4001.
Janssens GE, Meinema AC, Gonzalez J, Wolters JC, Schmidt A, Guryev V, et al. Protein biogenesis machinery is a driver of replicative aging in yeast. Elife. 2015;4: e08527. https://doi.org/10.7554/eLife.08527.
Berger AC, Vanderford TH, Gernert KM, Nichols JW, Faundez V, Corbett AH. Saccharomyces cerevisiae Npc2p is a functionally conserved homologue of the human Niemann-Pick disease type C 2 protein, hNPC2. Eukaryot Cell. 2005;4(11):1851–62. https://doi.org/10.1128/EC.4.11.1851-1862.2005.
Newton J, Milstien S, Spiegel S. Niemann-Pick type C disease: The atypical sphingolipidosis. Adv Biol Regul. 2018;70:82–8. https://doi.org/10.1016/j.jbior.2018.08.001.
Zhang S, Ren J, Li H, Zhang Q, Armstrong JS, Munn AL, et al. Ncr1p, the yeast ortholog of mammalian Niemann Pick C1 protein, is dispensable for endocytic transport. Traffic. 2004;5(12):1017–30. https://doi.org/10.1111/j.1600-0854.2004.00241.x.
Malathi K, Higaki K, Tinkelenberg AH, Balderes DA, Almanzar-Paramio D, Wilcox LJ, et al. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C-related protein reveals a primordial role in subcellular sphingolipid distribution. J Cell Biol. 2004;164(4):547–56. https://doi.org/10.1083/jcb.200310046.
Winkler MBL, Kidmose RT, Szomek M, Thaysen K, Rawson S, Muench SP, et al. Structural insight into eukaryotic sterol transport through Niemann-Pick type C proteins. Cell. 2019. https://doi.org/10.1016/j.cell.2019.08.038.
Hu Z, He B, Ma L, Sun Y, Niu Y, Zeng B. Recent advances in ergosterol biosynthesis and regulation mechanisms in Saccharomyces cerevisiae. Indian J Microbiol. 2017;57(3):270–7. https://doi.org/10.1007/s12088-017-0657-1.
Vambutas A, Ackerman SH, Tzagoloff A. Mitochondrial translational-initiation and elongation factors in Saccharomyces cerevisiae. Eur J Biochem. 1991;201(3):643–52.
Ott M, Amunts A, Brown A. Organization and regulation of mitochondrial protein synthesis. Annu Rev Biochem. 2016;85:77–101. https://doi.org/10.1146/annurev-biochem-060815-014334.
Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5(10): e261. https://doi.org/10.1371/journal.pbio.0050261.
Beach A, Leonov A, Arlia-Ciommo A, Svistkova V, Lutchman V, Titorenko VI. Mechanisms by which different functional states of mitochondria define yeast longevity. Int J Mol Sci. 2015;16(3):5528–54. https://doi.org/10.3390/ijms16035528.
Guaragnella N, Coyne LP, Chen XJ, Giannattasio S. (2018) Mitochondria-cytosol-nucleus crosstalk: learning from Saccharomyces cerevisiae. FEMS Yeast Res 18(8). doi:https://doi.org/10.1093/femsyr/foy088
Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–85. https://doi.org/10.1146/annurev.genet.40.110405.090613.
Zorova LD, Popkov VA, Plotnikov EY, Silachev DN, Pevzner IB, Jankauskas SS, et al. Mitochondrial membrane potential. Anal Biochem. 2018;552:50–9. https://doi.org/10.1016/j.ab.2017.07.009.
Jury DR, Kaveti S, Duan ZH, Willard B, Kinter M, Londraville R. Effects of calorie restriction on the zebrafish liver proteome. Comp Biochem Physiol Part D Genomics Proteomics. 2008;3(4):275–82. https://doi.org/10.1016/j.cbd.2008.07.003.
Valle A, Sastre-Serra J, Roca P, Oliver J. Modulation of white adipose tissue proteome by aging and calorie restriction. Aging Cell. 2010;9(5):882–94. https://doi.org/10.1111/j.1474-9726.2010.00613.x.
Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. https://doi.org/10.1146/annurev.cellbio.23.090506.123509.
Kim S, Villeponteau B, Jazwinski SM. Effect of replicative age on transcriptional silencing near telomeres in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1996;219(2):370–6. https://doi.org/10.1006/bbrc.1996.0240.
Konig J, Besoke F, Stuetz W, Malarski A, Jahreis G, Grune T, et al. Quantification of age-related changes of alpha-tocopherol in lysosomal membranes in murine tissues and human fibroblasts. BioFactors. 2016;42(3):307–15. https://doi.org/10.1002/biof.1274.
Demais V, Barthelemy A, Perraut M, Ungerer N, Keime C, Reibel S, et al. Reversal of pathologic lipid accumulation in NPC1-deficient neurons by drug-promoted release of LAMP1-coated lamellar inclusions. J Neurosci. 2016;36(30):8012–25. https://doi.org/10.1523/JNEUROSCI.0900-16.2016.
Wang YH, Twu YC, Wang CK, Lin FZ, Lee CY, Liao YJ. (2018) Niemann-Pick type C2 protein regulates free cholesterol accumulation and influences hepatic stellate cell proliferation and mitochondrial respiration function. Int J Mol Sci 19(6). doi:https://doi.org/10.3390/ijms19061678
Guo H, Zhao M, Qiu X, Deis JA, Huang H, Tang QQ, et al. Niemann-Pick type C2 deficiency impairs autophagy-lysosomal activity, mitochondrial function, and TLR signaling in adipocytes. J Lipid Res. 2016;57(9):1644–58. https://doi.org/10.1194/jlr.M066522.
Kennedy BE, Charman M, Karten B. Niemann-Pick Type C2 protein contributes to the transport of endosomal cholesterol to mitochondria without interacting with NPC1. J Lipid Res. 2012;53(12):2632–42. https://doi.org/10.1194/jlr.M029942.
Kennedy BE, Madreiter CT, Vishnu N, Malli R, Graier WF, Karten B. Adaptations of energy metabolism associated with increased levels of mitochondrial cholesterol in Niemann-Pick type C1-deficient cells. J Biol Chem. 2014;289(23):16278–89. https://doi.org/10.1074/jbc.M114.559914.
Solanko LM, Sullivan DP, Sere YY, Szomek M, Lunding A, Solanko KA, et al. Ergosterol is mainly located in the cytoplasmic leaflet of the yeast plasma membrane. Traffic. 2018;19(3):198–214. https://doi.org/10.1111/tra.12545.
Nielson JR, Fredrickson EK, Waller TC, Rendon OZ, Schubert HL, Lin Z et al. (2017) Sterol oxidation mediates stress-responsive Vms1 translocation to mitochondria. Mol Cell 68(4):673–85 e6. doi:https://doi.org/10.1016/j.molcel.2017.10.022
Altmann K, Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16(11):5410–7. https://doi.org/10.1091/mbc.e05-07-0678.
Krumpe K, Frumkin I, Herzig Y, Rimon N, Ozbalci C, Brugger B, et al. Ergosterol content specifies targeting of tail-anchored proteins to mitochondrial outer membranes. Mol Biol Cell. 2012;23(20):3927–35. https://doi.org/10.1091/mbc.E11-12-0994.
Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci U S A. 2008;105(40):15287–92. https://doi.org/10.1073/pnas.0807328105.
Vilaca R, Silva E, Nadais A, Teixeira V, Matmati N, Gaifem J, et al. Sphingolipid signalling mediates mitochondrial dysfunctions and reduced chronological lifespan in the yeast model of Niemann-Pick type C1. Mol Microbiol. 2014;91(3):438–51. https://doi.org/10.1111/mmi.12470.
Leupold S, Hubmann G, Litsios A, Meinema AC, Takhaveev V, Papagiannakis A et al. (2019) Saccharomyces cerevisiae goes through distinct metabolic phases during its replicative lifespan. Elife 8. doi:https://doi.org/10.7554/eLife.41046
Peric M, Bou Dib P, Dennerlein S, Musa M, Rudan M, Lovric A, et al. Crosstalk between cellular compartments protects against proteotoxicity and extends lifespan. Sci Rep. 2016;6:28751. https://doi.org/10.1038/srep28751.
Acknowledgements
We would like to thank Dr. Shelley Berger (UPenn) for her advice on this project. We also thank Dr. Michiel Vermeulen and Pascal Jansen (The Radboud University Medical Centre) for providing mass spectrometry analysis service.
Funding
This study was supported by NIH grants K99/R00AG037646, R56AG049714, R01AG052507, and R41/R42AG058368 to WD, CPRIT Scholar award R1306 to WD, Welch Foundation grant Q-1986–20190330 to WD, NSF award #1720215, #1761839, and internal support of the University of Tennessee at Chattanooga to HQ.
Author information
Authors and Affiliations
Contributions
Conceptualization, YS and WD; data curation, YS, RY, and WD; formal analysis, YS, RY, HG, HQ, and WD; funding acquisition, HQ, and WD; investigation, YS, RY, and HG; methodology, YS, RY, HG, and WD; project administration, YS and WD; resources, HQ, and WD; supervision, WD; validation, YS, RY and WD; writing — original draft, YS, RY and WD; writing — review and editing, YS, RY, HQ, and WD.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors have agreed to the content of this manuscript for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Sun, Y., Yu, R., Guo, HB. et al. A quantitative yeast aging proteomics analysis reveals novel aging regulators. GeroScience 43, 2573–2593 (2021). https://doi.org/10.1007/s11357-021-00412-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-021-00412-3