Abstract
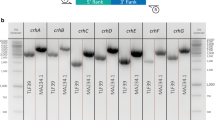
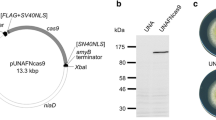
The CRISPR/Cas9 system has become a great tool for target gene knock-out in filamentous fungi. It is laborious and time-consuming that identification mutants from a large number of transformants through PCR or enzyme-cut method. Here, we first developed a CRISPR/Cas9 system in Aspergillus oryzae using AMA1-based autonomously replicating plasmid and Cas9 under the control of the Aspergillus nidulans gpdA promoter. By the genome editing technique, we successfully obtained mutations within each target gene in Aspergillus oryzae. Then, we put the protospacer sequence of a target gene and its protospacer adjacent motif (PAM) behind the start codon “ATG” of DsRed, yielding the non‑functional DsRed (nDsRed) reporter gene, and the nDsRed reporter gene could be rescued after successful targeted editing. Moreover, this method was also applied by targeting the kojic acid synthesis gene kojA, and the transformants with DsRed activity were found to harbor targeted mutations in kojA. These results suggest that the nDsRed can be used as a powerful tool to facilitate the identification of mutants generated by CRISPR/Cas9 in Aspergillus oryzae.





Similar content being viewed by others
References
Chutrakul C, Panchanawaporn S, Jeennor S, Anantayanon J, Vorapreeda T, Vichai V, Laoteng K (2019) Functional characterization of novel U6 RNA polymerase III promoters: their implication for CRISPR-Cas9-mediated gene editing in Aspergillus oryzae. Curr Microbiol 76:1443–1451. https://doi.org/10.1007/s00284-019-01770-0
Doench JG et al (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32:1262–1267. https://doi.org/10.1038/nbt.3026
Donohoue PD, Barrangou R, May AP (2018) Advances in industrial biotechnology using CRISPR-Cas systems. Trends Biotechnol 36:134–146. https://doi.org/10.1016/j.tibtech.2017.07.007
Fan J, Zhang Z, Long C, He B, Hu Z, Jiang C, Zeng B (2020) Identification and functional characterization of glycerol dehydrogenase reveal the role in kojic acid synthesis in Aspergillus oryzae. World J Microbiol Biotechnol. https://doi.org/10.1007/s11274-020-02912-4
Haeussler M et al (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17:148. https://doi.org/10.1186/s13059-016-1012-2
Hsu PD et al (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832. https://doi.org/10.1038/nbt.2647
Jiang F, Doudna JA (2017) CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529. https://doi.org/10.1146/annurev-biophys-062215010822
Kadooka C et al (2020) A CRISPR/Cas9-mediated gene knockout system in Aspergillus luchuensis mut. kawachii. Biosci Biotechnol Biochem 84:2179–2183. https://doi.org/10.1080/09168451.2020.1792761
Katayama T, Nakamura H, Zhang Y, Pascal A, Fujii W, Maruyama JI (2019) Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus Aspergillus oryzae. Appl Environ Microbiol 85:e01896-e1918. https://doi.org/10.1128/AEM.01896-18
Katayama T, Tanaka Y, Okabe T, Nakamura H, Fujii W, Kitamoto K, Maruyama J (2016) Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol Lett 38:637–642. https://doi.org/10.1007/s10529-015-2015-x
Kobayashi T et al (2007) Genomics of Aspergillus oryzae. Biosci Biotechnol Biochem 71:646–670. https://doi.org/10.1271/bbb.60550
Labuhn M et al (2018) Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res 46:1375–1385. https://doi.org/10.1093/nar/gkx1268
Lander ES (2016) The heroes of CRISPR. Cell 164:18–28. https://doi.org/10.1016/j.cell.2015.12.041
Ma X et al (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284. https://doi.org/10.1016/j.molp.2015.04.007
Machida M et al (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161. https://doi.org/10.1038/nature04300
Malina A, Cameron CJF, Robert F, Blanchette M, Dostie J, Pelletier J (2015) PAM multiplicity marks genomic target sites as inhibitory to CRISPR-Cas9 editing. Nat Commun 6:10124. https://doi.org/10.1038/ncomms10124
Maruyama J, Kitamoto K (2011) Targeted gene disruption in Koji mold Aspergillus oryzae. Methods Mol Biol 765:447–456. https://doi.org/10.1007/978-1-61779-197-0_27
Nguyen KT et al (2017) A new and efficient approach for construction of uridine/uracil auxotrophic mutants in the filamentous fungus Aspergillus oryzae using Agrobacterium tumefaciens-mediated transformation. World J Microbiol Biotechnol 33:107. https://doi.org/10.1007/s11274-017-2275-9
Nodvig CS, Hoof JB, Kogle ME, Jarczynska ZD, Lehmbeck J, Klitgaard DK, Mortensen UH (2018) Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet Biol 115:78–89. https://doi.org/10.1016/j.fgb.2018.01.004
Nodvig CS, Nielsen JB, Kogle ME, Mortensen UH (2015) A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 10:e0133085. https://doi.org/10.1371/journal.pone.0133085
Osakabe Y et al (2018) CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat Protoc 13:2844–2863. https://doi.org/10.1038/s41596-018-0067-9
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9:2395–2410. https://doi.org/10.1038/nprot.2014.157
Song L, Ouedraogo JP, Kolbusz M, Nguyen TTM, Tsang A (2018) Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger. PLoS ONE 13:e0202868. https://doi.org/10.1371/journal.pone.0202868
Song R et al (2019) CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Appl Microbiol Biotechnol 103:6919–6932. https://doi.org/10.1007/s00253-019-10007-w
Terabayashi Y et al (2010) Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet Biol 47:953–961. https://doi.org/10.1016/j.fgb.2010.08.014
Tsai SQ et al (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33:187–197. https://doi.org/10.1038/nbt.3117
Vouillot L, Thelie A, Pollet N (2015) Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 5:407–415. https://doi.org/10.1534/g3.114.015834
Wang Q, Coleman JJ (2019) Progress and challenges: development and implementation of CRISPR/Cas9 technology in filamentous fungi. Comput Struct Biotechnol J 17:761–769. https://doi.org/10.1016/j.csbj.2019.06.007
Weyda I, Yang L, Vang J, Ahring BK, Lubeck M, Lubeck PS (2017) A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J Microbiol Methods 135:26–34. https://doi.org/10.1016/j.mimet.2017.01.015
Zhang C, Meng X, Wei X, Lu L (2016) Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet Biol 86:47–57. https://doi.org/10.1016/j.fgb.2015.12.007
Acknowledgements
We thank Dr. Van-Tuan Tran for kindly providing the plasmids pEX1 and pEX2B. This work was supported by Natural Science Foundation of China (32000049), Jiangxi Provincial Natural Science Foundation (20202BABL213042), Science and Technology Research Project of Jiangxi Provincial Department of Education (GJJ190611), and Doctoral Scientific Research Foundation of Jiangxi Science and Technology Normal University (2018BSQD030).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals experiments.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Zhang, H., Fan, J. et al. A highly efficient identification of mutants generated by CRISPR/Cas9 using the non‑functional DsRed assisted selection in Aspergillus oryzae. World J Microbiol Biotechnol 37, 132 (2021). https://doi.org/10.1007/s11274-021-03100-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-021-03100-8