Abstract
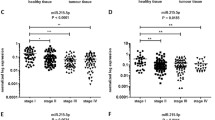
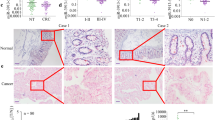
Colorectal carcinoma (CRC) results from the accumulation of genetic mutations and alterations in signaling pathways. KRAS is mutated in 40% of CRC cases and is involved in increased tumor cells proliferation and survival. Although KRAS mutations are a dominant event in CRC tumorigenesis, increased wild-type KRAS expression has a similar effect on accelerated tumor growth. In this study, we investigated the KRAS status in correlation with clinicopathological features in sporadic CRC and more importantly the role of let-7a-5p and miR-544a-3p in the regulation of wild-type KRAS protein expression in the tumor center (T1) and invasive tumor front (T2). Analysis showed that 39.1% of tumor samples had KRAS mutations. In wild-type KRAS tumors, 62.0% were positive for KRAS protein expression and there was a higher percentage of KRAS-positive tumor cells and a higher intensity of immunohistochemical reaction in T2 than in T1 samples. This could not be attributed to differences in KRAS mRNA levels, suggesting regulation via miR-544a-3p expression which was significantly decreased in T2 samples. Furthermore, we demonstrated that tumor samples carrying the KRAS-LCS6 variant allele had significantly higher protein expression of the wild-type KRAS. Our results suggest the role of the KRAS-LCS6 polymorphism and miR-544a-3p expression in the regulation of wild-type KRAS protein expression in sporadic CRC.





Similar content being viewed by others
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Mondaca S, Yaeger R. Colorectal cancer genomics and designing rational trials. Ann Transl Med. 2018;6(9):159.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49.
Tsilimigras DI, Ntanasis-Stathopoulos I, Bagante F, Moris D, Cloyd J, Spartalis E, et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: a systematic review of the current evidence. Surg Oncol. 2018;27(2):280–8.
Guerrero S, Casanova I, Farre L, Mazo A, Capella G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60(23):6750–6.
Horsch M, Recktenwald CV, Schadler S, Hrabe de Angelis M, Seliger B, Beckers J. Overexpressed vs mutated Kras in murine fibroblasts: a molecular phenotyping study. Br J Cancer. 2009;100(4):656–62.
Zhou B, Der CJ, Cox AD. The role of wild type RAS isoforms in cancer. Semin Cell Dev Biol. 2016;58:60–9.
Roncarati R, Lupini L, Shankaraiah RC, Negrini M. The importance of microRNAs in RAS oncogenic activation in human cancer. Front Oncol. 2019;9:988.
You C, Liang H, Sun W, Li J, Liu Y, Fan Q, et al. Deregulation of the miR-16-KRAS axis promotes colorectal cancer. Sci Rep. 2016;6:37459.
Kim M, Slack FJ. MicroRNA-mediated regulation of KRAS in cancer. J Hematol Oncol. 2014;7:84.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47.
Ma R, Zhang G, Wang H, Lv H, Fang F, Kang X. Downregulation of miR-544 in tissue, but not in serum, is a novel biomarker of malignant transformation in glioma. Oncol Lett. 2012;4(6):1321–4.
Yanaka Y, Muramatsu T, Uetake H, Kozaki K, Inazawa J. miR-544a induces epithelial-mesenchymal transition through the activation of WNT signaling pathway in gastric cancer. Carcinogenesis. 2015;36(11):1363–71.
Liang D, Meyer L, Chang DW, Lin J, Pu X, Ye Y, et al. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Can Res. 2010;70(23):9765–76.
Gottmann P, Ouni M, Zellner L, Jahnert M, Rittig K, Walther D, et al. Polymorphisms in miRNA binding sites involved in metabolic diseases in mice and humans. Sci Rep. 2020;10(1):7202.
Jacinta-Fernandes A, Xavier JM, Magno R, Lage JG, Maia AT. Allele-specific miRNA-binding analysis identifies candidate target genes for breast cancer risk. NPJ Genom Med. 2020;5:4.
Gholami M, Larijani B, Sharifi F, Hasani-Ranjbar S, Taslimi R, Bastami M, et al. MicroRNA-binding site polymorphisms and risk of colorectal cancer: a systematic review and meta-analysis. Cancer Med. 2019;8(17):7477–99.
Sclafani F, Chau I, Cunningham D, Peckitt C, Lampis A, Hahne JC, et al. Prognostic role of the LCS6 KRAS variant in locally advanced rectal cancer: results of the EXPERT-C trial. Ann Oncol. 2015;26(9):1936–41.
Ryan BM, Robles AI, Harris CC. KRAS-LCS6 genotype as a prognostic marker in early-stage CRC–letter. Clin Cancer Res. 2012;18(12):3487–8.
Usher-Smith JA, Walter FM, Emery JD, Win AK, Griffin SJ. Risk prediction models for colorectal cancer: a systematic review. Cancer Prev Res (Phila). 2016;9(1):13–26.
Dou R, Nishihara R, Cao Y, Hamada T, Mima K, Masuda A, et al. MicroRNA let-7, T cells, and patient survival in colorectal cancer. Cancer Immunol Res. 2016;4(11):927–35.
Luu C, Heinrich EL, Duldulao M, Arrington AK, Fakih M, Garcia-Aguilar J, et al. TP53 and let-7a micro-RNA regulate K-Ras activity in HCT116 colorectal cancer cells. PLoS ONE. 2013;8(8):70604.
Sha D, Lee AM, Shi Q, Alberts SR, Sargent DJ, Sinicrope FA, et al. Association study of the let-7 miRNA-complementary site variant in the 3’ untranslated region of the KRAS gene in stage III colon cancer (NCCTG N0147 Clinical Trial). Clin Cancer Res. 2014;20(12):3319–27.
Fang C, Li Y. Prospective applications of microRNAs in oral cancer. Oncol Lett. 2019;18(4):3974–84.
Ghosh RD, Pattatheyil A, Roychoudhury S. Functional landscape of dysregulated microRNAs in oral squamous cell carcinoma: clinical implications. Front Oncol. 2020;10:619.
Paterson EL, Kazenwadel J, Bert AG, Khew-Goodall Y, Ruszkiewicz A, Goodall GJ. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia. 2013;15(2):180–91.
Spaventi R, Pecur L, Pavelic K, Pavelic ZP, Spaventi S, Stambrook PJ. Human tumour bank in Croatia: a possible model for a small bank as part of the future European tumour bank network. Eur J Cancer. 1994;30A(3):419.
Nollau P, Moser C, Weinland G, Wagener C. Detection of K-ras mutations in stools of patients with colorectal cancer by mutant-enriched PCR. Int J Cancer. 1996;66(3):332–6.
Saridaki Z, Weidhaas JB, Lenz HJ, Laurent-Puig P, Jacobs B, De Schutter J, et al. A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy. Clin Cancer Res. 2014;20(17):4499–510.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8.
D’Agostino RB, Belanger A, D’Agostino RB Jr. A suggestion for using powerful and informative test of normality. Am Stat. 1990;44(4):316–21.
Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–99.
Crespo P, Leon J. Ras proteins in the control of the cell cycle and cell differentiation. Cell Mol Life Sci. 2000;57(11):1613–36.
Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5(4):375–87.
Valtorta E, Misale S, Sartore-Bianchi A, Nagtegaal ID, Paraf F, Lauricella C, et al. KRAS gene amplification in colorectal cancer and impact on response to EGFR-targeted therapy. Int J Cancer. 2013;133(5):1259–65.
Serebriiskii IG, Connelly C, Frampton G, Newberg J, Cooke M, Miller V, et al. Comprehensive characterization of RAS mutations in colon and rectal cancers in old and young patients. Nat Commun. 2019;10(1):3722.
Guo F, Gong H, Zhao H, Chen J, Zhang Y, Zhang L, et al. Mutation status and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese colorectal cancer patients. Sci Rep. 2018;8(1):6076.
Chuang SC, Huang CW, Chen YT, Ma CJ, Tsai HL, Chang TK, et al. Effect of KRAS and NRAS mutations on the prognosis of patients with synchronous metastatic colorectal cancer presenting with liver-only and lung-only metastases. Oncol Lett. 2020;20(3):2119–30.
Liu P, Wang Y, Li X. Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B. 2019;9(5):871–9.
Piton N, Lonchamp E, Nowak F, Sabourin JC. Real-life distribution of KRAS and NRAS mutations in metastatic colorectal carcinoma from French routine genotyping. Cancer Epidemiol Biomarkers Prev. 2015;24(9):1416–8.
van Wyk HC, Roseweir A, Alexander P, Park JH, Horgan PG, McMillan DC, et al. The relationship between tumor budding, tumor microenvironment, and survival in patients with primary operable colorectal cancer. Ann Surg Oncol. 2019;26(13):4397–404.
Ozer SP, Barut SG, Ozer B, Catal O, Sit M. The relationship between tumor budding and survival in colorectal carcinomas. Rev Assoc Med Bras (1992). 2019;65(12):1442–7.
Dawson H, Lugli A. Molecular and pathogenetic aspects of tumor budding in colorectal cancer. Front Med (Lausanne). 2015;2:11.
Trinh A, Ladrach C, Dawson HE, Ten Hoorn S, Kuppen PJK, Reimers MS, et al. Tumour budding is associated with the mesenchymal colon cancer subtype and RAS/RAF mutations: a study of 1320 colorectal cancers with Consensus Molecular Subgroup (CMS) data. Br J Cancer. 2018;119(10):1244–51.
Maffeis V, Nicole L, Cappellesso R. RAS, cellular plasticity, and tumor budding in colorectal cancer. Front Oncol. 2019;9:1255.
Rimbert J, Tachon G, Junca A, Villalva C, Karayan-Tapon L, Tougeron D. Association between clinicopathological characteristics and RAS mutation in colorectal cancer. Mod Pathol. 2018;31(3):517–26.
Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol. 2013;26(6):825–34.
Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–6.
Elsabah MT, Adel I. Immunohistochemical assay for detection of K-ras protein expression in metastatic colorectal cancer. J Egypt Natl Canc Inst. 2013;25(1):51–6.
Piton N, Borrini F, Bolognese A, Lamy A, Sabourin JC. KRAS and BRAF Mutation Detection: Is Immunohistochemistry a Possible Alternative to Molecular Biology in Colorectal Cancer? Gastroenterol Res Pract. 2015;2015:753903.
Dhawan A, Scott JG, Harris AL, Buffa FM. Pan-cancer characterisation of microRNA across cancer hallmarks reveals microRNA-mediated downregulation of tumour suppressors. Nat Commun. 2018;9(1):5228.
Niveditha D, Jasoria M, Narayan J, Majumder S, Mukherjee S, Chowdhury R, et al. Common and unique microRNAs in multiple carcinomas regulate similar network of pathways to mediate cancer progression. Sci Rep. 2020;10(1):2331.
Jevsinek Skok D, Hauptman N, Bostjancic E, Zidar N. The integrative knowledge base for miRNA-mRNA expression in colorectal cancer. Sci Rep. 2019;9(1):18065.
Fadaka AO, Pretorius A, Klein A. MicroRNA assisted gene regulation in colorectal cancer. Int J Mol Sci. 2019;20(19):4899.
Moszynska A, Gebert M, Collawn JF, Bartoszewski R. SNPs in microRNA target sites and their potential role in human disease. Open Biol. 2017;7(4):170019.
Chin LJ, Ratner E, Leng SG, Zhai RH, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Can Res. 2008;68(20):8535–40.
Chen M, Liu YY, Zheng MQ, Wang XL, Gao XH, Chen L, et al. microRNA-544 promoted human osteosarcoma cell proliferation by downregulating AXIN2 expression. Oncol Lett. 2018;15(5):7076–82.
Smits KM, Paranjape T, Nallur S, Wouters KA, Weijenberg MP, Schouten LJ, et al. A let-7 microRNA SNP in the KRAS 3’UTR is prognostic in early-stage colorectal cancer. Clin Cancer Res. 2011;17(24):7723–31.
Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, et al. A let-7 microRNA-binding site polymorphism in the KRAS 3’ UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30(6):1003–7.
Liang D, Meyer L, Chang DW, Lin J, Pu X, Ye Y, et al. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70(23):9765–76.
Ganzinelli M, Rulli E, Caiola E, Garassino MC, Broggini M, Copreni E, et al. Role of KRAS-LCS6 polymorphism in advanced NSCLC patients treated with erlotinib or docetaxel in second line treatment (TAILOR). Sci Rep. 2015;5:16331.
Nelson HH, Christensen BC, Plaza SL, Wiencke JK, Marsit CJ, Kelsey KT. KRAS mutation, KRAS-LCS6 polymorphism, and non-small cell lung cancer. Lung Cancer. 2010;69(1):51–3.
Uvirova M, Simova J, Kubova B, Dvorackova N, Tomaskova H, Sedivcova M, et al. Comparison of the prevalence of KRAS-LCS6 polymorphism (rs61764370) within different tumour types (colorectal, breast, non-small cell lung cancer and brain tumours). A study of the Czech population. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2015;159(3):466–71.
Funding
The present study was supported by the Croatian Science Foundation (Grant number HRZZ-IP-2016-06-1430).
Author information
Authors and Affiliations
Contributions
SM: investigation, formal analysis, and writing—original draft, AŠ: resources, formal analysis, and visualization, TCI: investigation and formal analysis, MP: resources, SK: conceptualization, funding acquisition, and writing—reviewing and editing.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval and informed consent to participate
Written informed consent was obtained from all patients included in the study. The study was approved by the ethics committee of Merkur Clinical Hospital, Zagreb and Medical School, University of Zagreb, and was performed under the ethical standards of the Helsinki Declaration.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Marinović, S., Škrtić, A., Catela Ivković, T. et al. Regulation of KRAS protein expression by miR-544a and KRAS-LCS6 polymorphism in wild-type KRAS sporadic colon adenocarcinoma. Human Cell 34, 1455–1465 (2021). https://doi.org/10.1007/s13577-021-00576-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00576-2