Abstract
The syntheses of [Cu(PPh3)2(L)]NO3 and [Cu(PPh3)2(L-SO3Na)]NO3 were achieved through the reaction of Cu(PPh3)2NO3 and equimolar amount of the ligands (L = 5,6-diphenyl-3-[2-pyridyl]-1,2,4-triazine; LSO3Na = 5,6-diphenyl-3-[2-pyridyl]-1,2,4-triazine-4,4′-disulfonic acid disodium salt). The complexes were characterized by NMR and IR spectroscopy and mass spectrometry. The compounds exhibit similar absorption and emission spectra, suggesting a similar electronic structure. Ct-DNA binding studies show the strong influence of the net charge as Cu-L (positively charged) is able to bind to ct-DNA while Cu-LSO3Na (negatively charged) is not. The net charge of the complexes affects the thermodynamic and kinetic binding parameters toward human serum albumin. HSA-binding of the complexes was further investigated by molecular docking, revealing different binding sites on the HSA protein as a function of the net charge. The different anticancer activities of the complexes towards ovcar-3 and hope-62 cancer cell lines are suggestive of a role for the overall charge of the complexes. Interaction with the DNA is not the major mechanism for this class of complexes. The overall net charge of the pharmacophore (anticancer agent) should be a key consideration in the design of anticancer metal complexes.
Similar content being viewed by others
1 Introduction
Several transition-metal complexes, mainly containing platinum, have been approved or are currently in clinical trials for chemotherapy. Cisplatin is one of the most widespread chemotherapeutic drugs, being used in 50% of the chemotherapeutic treatment regimes, but its severe side effects have motivated further research efforts to develop alternatives [1,2,3]. Carboplatin and oxaliplatin have been approved (by the FDA) for clinical use while nedaplatin, lobaplatin, and heptaplatin have gained approval for clinical use [4]. Another strategy for the design of chemotherapeutic agents has involved the use of different metals, such as ruthenium, palladium and gold [5,6,7]. Promising coordination compounds of copper(I) and copper(II) have been examined as candidates in cancer therapy, aiming for less toxic compounds than platinum compounds with comparable or better activities (because copper is an essential trace element in biological systems) [8,9,10]. In particular, the interest in copper(I) complexes with mixed phosphine/diimine ligand sets has been growing tremendously. Recent research has confirmed the role of the diimine ligands in improving the cytotoxic properties of the copper complexes. A set of copper(I) complexes of the type [CuX(N^N)(tris-(2-cyanoethyl)phosphine)] has been synthesized and evaluated for their in vitro antineoplastic properties against several cancer cell lines, highlighting that the most effective complex was the one with a dipyrido-[3,2-d:2′,3′-f]-quinoxaline ligand [11]. In another study, a set of complexes with the general formula [CuBr(PPh3)(N^N)] was examined for their anticancer activities, the authors concluding that complexes with 3-[2-pyridyl]-5,6-diphenyl-1,2,4-triazine and dipyrido[3,2-a:2′,3′-c]phenazine (dppz) exhibited better cytotoxicity against several cancer cell lines than complexes with substituted phenanthroline and bipyridines [12]. Moreover, functionalizing the dppz ligand with groups such as nitro, cyano, methyl, etc., has been shown to alter the anticancer properties significantly, proving the critical role of the diimine ligands in the biological properties [13]. Other reports have examined the effect of incorporating hydrophilic phosphines such as tris(hydroxymethyl)phosphine [14, 15] and N-methyl-1,3,5-triaza-7-phosphaadamantane [16]; these limited studies have concluded that the water-soluble complexes have good anticancer activities with improved physiochemical properties. Copper(I) complexes including diimine ligands with water-solubilizing groups have also been examined for their in vitro anticancer potential against human tumor cell lines, exhibiting moderate to high cytotoxic activities and with potential to overcome cisplatin resistant cell lines [17]. Other strategies employed to improve the delivery of chemotherapy include functionalization of the drug with ester or carboxylic acid moieties [18,19,20], assembling pH- or temperature-responsive multilayer polymers encapsulating drugs [21,22,23], and encapsulating drugs in nanosheets [24] or metal–organic frameworks [25].
As mentioned above, the use of 5,6-diphenyl-3-[2-pyridyl]-1,2,4-triazine (L) as a ligand in CuBr(PPh3)(L) improves the anticancer activities. In this work, water-solubilizing sodium sulfonate units were incorporated into the 5,6-diphenyl-3-[2-pyridyl]-1,2,4-triazine ligand (affording L-SO3Na). Two complexes were synthesized with the formula [Cu(PPh3)2(L)]NO3 and [Cu(PPh3)2(L-SO3Na)]NO3, and the DNA-binding, protein-binding and anticancer activities were evaluated to assess the role of the complex net charge.
2 Experimental Section
2.1 Chemicals and Reagents
Solvents (HPLC grade) were used as received. Calf thymus DNA (Type I) and human serum albumin were purchased from Sigma-Aldrich. 5,6-Diphenyl-3-(2-pyridyl)-1,2,4-triazine (L) and 5,6-diphenyl-3-(2-pyridyl)-1,2,4-triazine-4,4′-disulfonic acid disodium salt hydrate (L-SO3Na) were purchased from Alfa Aesar. The synthesis of Cu(PPh3)2NO3 was achieved as described in the literature, by stirring copper(II) nitrate hemi(pentahydrate) with ca. three molar equivalents of triphenylphosphine in ethanol at reflux under a nitrogen gas atmosphere. The desired compound precipitated after a few minutes and was obtained by filtration as a white powder [26].
2.2 Instrumentation
Infrared spectra were recorded for powder samples of the compounds, using a Bruker Alpha FT-IR; peaks are reported in cm−1. 1H NMR (600 MHz) and 31P NMR (242 MHz) spectra were recorded using a Bruker Avance 600 MHz spectrometer equipped with a BBO probe (BrukerBioSpin, Rheinstetten, Germany). The spectra were recorded using previously reported parameters [27]. High-resolution electrospray ionization (ESI) mass spectra were recorded using an Agilent Q-TOF 6520 instrument; all mass spectrometry data are reported as m/z. Absorption spectroscopy was performed using a Genesys-10 s UV–VIS spectrophotometer and 1 cm path-length quartz cells; bands are reported in the form wavelength (nm). Emission spectra were collected in 1 cm quartz cells using a Hitachi F-7000 fluorescence spectrometer.
2.2.1 Synthesis of [Cu(PPh3)2(L)] (Cu-L)
A mixture of Cu(PPh3)2NO3 (600 mg, 0.922 mmol) and 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine (286 mg, 0.922 mmol) in dichloromethane (15 mL) was stirred for 2 h under a nitrogen atmosphere. The reaction solution was reduced in volume, diethyl ether was added, and the orange precipitate was collected by filtration, washed with petrol, and dried, affording complex Cu-L as an orange powder (420 mg, 51%). IR (solid, cm−1): 3050 w, 1597 w, 1509 w, 1479 m, 1434 m, 1410 w, 1342 br, 1182 w, 1158 w, 1095 m, 1025 w, 997 w, 829 w, 797 w, 7706 m, 769 s, 744 s, 602 s, 644 w, 618 w. 1H NMR (CDCl3, 600 MHz, 298 K): δ 8.79 (s br, 1H, py), 8.50 (s br, 1H, py), 8.31 (s br, 1H, py), 7.78 (s br, 1H, py), 7.62 (d, JHH = 6 Hz, 2H, Ph), 7.52 (s br, 2H, Ph), 7.44 (s br, 4H, Ph), 7.31 (s br, 6H, p-Ph PPh3), 7.16 (s br, 24H, PPh3). 31P {1H} NMR (CDCl3, 600 MHz, 298 K): 3.17 (s, PPh3). HR ESI MS: calcd for [C38H29CuN4P] + [M-PPh3] + 635.1425, found 635.14201.
2.2.2 Synthesis of [Cu(PPh3)2(L-SO3Na)] (Cu-LSO3Na)
Cu(PPh3)2NO3 (503 mg, 770 mmol) and LSO3Na (0.398 g, 770 mmol) were stirred in a mixture of acetonitrile (10 mL) and methanol (5 mL) with a few drops of water for 2 h under a nitrogen atmosphere. The reaction solution was reduced in volume, diethyl ether was added, and the orange precipitate was collected by filtration, washed with petrol, and dried, affording complex Cu-LSO3Na as an orange powder (262 mg, 31%). IR (solid, cm−1): 3459 br, 3055 w, 1638 m, 1597 w, 1511w, 1480 m, 1435 m, 1393 w, 1365 m, 1196 br, 1156 m, 1117 m, 1096 m, 1033 s, 996 w, 907 w, 800 m, 784 m, 746 s, 740 s, 650 w, 617 s, 574 w, 561 w. 1H NMR (CDCl3, 600 MHz, 298 K): δ 8.76 (d, JHH = 12 Hz, 1H, py), 8.34 (d, JHH = 6 Hz, 2H, Ph), 8.14 (t, JHH = 7 Hz, 1H, py), 8.07 (s br, 1H, py), 7.99 (s br, 2H, Ph), 7.53 (t, JHH = 7 Hz, 1H, py), 7.26 (s br, 4H, Ph), 7.19 (s br, 6H, p-Ph PPh3), 7.06 (s br, 24H, PPh3). 31P {1H} NMR (CDCl3, 600 MHz, 298 K): 3.21 (s, PPh3). HR ESI MS: calcd for [C38H27CuN4O6PS2] + [M-2Na-PPh3] + 839.02011, found 839.0287.
2.3 DNA Binding Studies
2.3.1 Ethidium Bromide Fluorescence Quenching
A solution containing 100 µM ct-DNA and 10 µM ethidium bromide (EtBr) was prepared in Tris–HCl/NaCl buffer solution (pH = 7.2). The ct-DNA-EtBr solution was incubated for 24 h. The solution was then titrated with different amounts of each of the complexes in DMSO, maintaining the DMSO at ca. 5% V/V. The changes in the emission spectra of the solutions were followed in the range 500–700 nm upon excitation at 390 nm after incubating the complexes for 4 min. The Stern–Volmer quenching constants (KSV) were calculated employing the equation below:
where Fo and F are the emission intensities in the absence and the presence of the samples, respectively. The [Cu compound] was plotted against [Fo/F]; the KSV value was equal to the slope [28, 29].
2.3.2 Determination of Binding Mode by Viscometry
An Ostwald viscometer was used to perform the viscosity measurements. 10 µL amounts of copper complexes were added to solutions of ct-DNA in buffer during which the [Cu compound]/[DNA] ratio was maintained in the range 0.02 to 0.2. The solutions were incubated for 5 min at 25 °C in a water bath before measurements. Average time flow for the solutions were recorded (replicating the measurements four times). The relative viscosities (η /ηo)1/3 were plotted against [Cu compound]/[DNA], where ηo and η represent the specific viscosity of the ct-DNA and the ct-DNA-Cu compound mixture, respectively. The specific viscosity η and ηo were obtained utilizing the relation [(t−tb)/tb] where t is the observed flow time and tb is the buffer solution flow time [28, 29].
2.4 HSA Binding Studies
A solution containing 30 µM of human serum albumin (HSA) was prepared in Tris–HCl/NaCl aqueous buffer system (pH = 7.4). The solution was titrated with different concentrations of both complexes in DMSO (the amount of DMSO was maintained at ca. 20% V/V). The changes in the emission spectra of the solutions were monitored at around 330 nm upon excitation at 270 nm, after incubating the complexes for 4 min at 295 K. The Stern–Volmer quenching constants (KSV) were calculated using Eq. 1 where Fo and F are the emission intensities of the protein in the absence and the presence of the complexes, respectively. The [Cu compound] was plotted against [Fo/F]; the KSV value was equal to the slope [30].
Binding constant (Kb, M−1), and the number of binding sites (n) were calculated by plotting log[Fo−F/F] against log [Cu compounds] where the slope equals n and the intercept equals log Kb, as can be expressed in Eq. 2 [31].
The binding constant (Kb, M−1) can be used to calculate the change in Gibbs free energy (∆G0) from the following equation [31, 32]:
2.5 Molecular Docking
The structure of the HSA receptor was downloaded from the protein data bank (PDB: 1H9Z) (http://www.rcsb.org/pdb/home/home.do) [33]. The two synthesized compounds were sketched in ChemBioOffice Ultra, version 13. All docking studies used the MOE program. HSA (1H9Z) was prepared by removing all water and cofactor molecules from the downloaded proteins. Then, all invalid charges and broken bonds were corrected, and all hydrogen atoms were added. The parameters and charges were assigned with the MMFF94x force field. After alpha-site spheres were identified using the site finder module of MOE, the two complexes were docked to the same active site of the downloaded compound using the DOCK module of MOE [34, 35]. The docking scores in MOE software were obtained using the London dG scoring function. The highest ten docking scores were used to compare between the two complexes and the co-crystallized reference compound; the values were optimized using two independent refinement methods. The docking results were validated following the reported method [36, 37]. The method (the pose selection method) involves re-docking the co-crystallized ligand into the receptor's active site. If the program is able to identify the best pose under a preselected Root Mean Square Deviation (RMSD) value from the known conformation (regularly 1.5 or 2 Å, depending on ligand size), the method is considered “validated”. In the current study, pose selection and docking score for R-warfarin (the co-crystallized compound with 1H9Z) were determined; the docking result of the same compound reached a 1.09 Å resolution of the co-crystal structure.
2.6 Anticancer Studies
The cells were supplied by the Egyptian Holding Company for Biological Products and Vaccines (VACSERA) and then kept in the tissue culture unit. The growth of the cells was effected in RPMI‐1640 medium, supplemented with 10% heat inactivated FBS, 50 units/mL of penicillin, and 50 mg/mL of streptomycin, and maintained in a humidified atmosphere with 5% carbon dioxide [38, 39]. The cells were maintained as monolayer cultures by serial sub‐culturing, with cell culture reagents obtained from Lonza (Basel, Switzerland). The antitumor activities of the complexes were assessed against OVCAR-3 (ovarian cancer) and HOPE-62 (small cell lung cancer) cell lines.
The sulforhodamine B (SRB) assay method was applied to determine the cytotoxicity, as described in the literature [40]. Exponentially-growing cells were collected using 0.25% Trypsin‐EDTA and seeded in 96‐well plates at 1000–2000 cells/well in RBMI‐1640 supplemented medium. The cells were kept in the medium for 24 h and then they were incubated for 3 days with various concentrations of the copper complexes. Following 3 days of treatment, the cells were fixed with 10% trichloroethanoic acid for 1 h at 4 °C. Wells were stained for 10 min at room temperature with 0.4% SRBC dissolved in 1% acetic acid. The plates were air dried for 24 h and the dye was dissolved in Tris‐HCl for 5 min with shaking at 1600 rpm. The optical density (OD) of each well was assessed spectrophotometrically at 564 nm with an ELISA microplate reader (ChroMate‐4300, FL, USA). The IC50 values were calculated from a Boltzmann sigmoidal concentration response curve using the nonlinear regression fitting models (Graph Pad, Prism Version 9).
3 Results and Discussion
3.1 Synthesis and Characterization
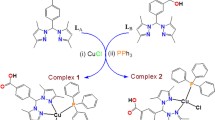
An initial proposal was made to compare CuBr(PPh3)(L) against CuBr(PPh3)(L-SO3Na), but several problems were encountered in the synthesis of pure compound of the latter, including the existence of the ligand in two different coordination modes. Hence, we shifted our work to the synthesis of complexes of the formula [Cu(PPh3)2(N^N)]NO3. Copper(I) complexes were prepared as described in Fig. 1, by reacting stoichiometric amounts of the ligands and Cu(PPh3)2NO3 under an inert gas atmosphere; the complexes were obtained in moderate yields. For the ligand 5,6-diphenyl-3-(2-pyridyl)-1,2,4-triazine-4,4′-disulfonic acid disodium salt hydrate (L-SO3Na), the use of water was essential to dissolve the ligand, and hence we used acetonitrile/methanol mixture with a few drops of water to run the reaction. For 5,6-diphenyl-3-(2-pyridyl)-1,2,4-triazine, the reaction was performed in dichloromethane.
Both complexes reported in this work are stable at ambient conditions as solids or as solutions in chlorinated solvents or DMSO. The identities of the complexes were confirmed by 1H and 31P NMR spectroscopies as well as elemental analysis and mass spectrometry. In the 31P NMR spectra, the compounds show intense singlet peaks around − 6 ppm. The mass spectra provided signals for [M-PPh3-NO3]+ in the case of Cu-L while the ion [M-PPh3-2Na-NO3]+ was detected for Cu-LSO3Na. The IR spectrum of Cu-LSO3Na confirms the presence of nitrate ion with the signal at 1365 cm−1. Electronic spectra of both compounds are almost identical, as both compounds exhibit an intense band at around 280 nm with a shoulder extending to ca. 370 nm (Fig. 2). The intense high energy bands are assigned as LC (π–π*) and the broad shoulder bands are attributed to MLCT (d-π*) [12, 13, 41]. The emission spectra of the complexes were measured in deoxygenated DMSO solutions, with both compounds being excited at 370 nm; the excitation wavelength was chosen experimentally by scanning in the proximity of the bands in the absorption spectra. The compounds have similar emission behavior with emission maxima at 420 nm and 450 nm, with a shoulder extending to 550 nm (Fig. 2).
3.2 DNA Binding Interactions
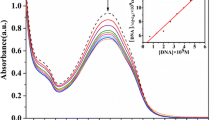
Ethidium bromide is weakly emissive, the emission being enhanced when EtBr intercalates between the base pairs in the DNA, so the emissive EtBr-DNA adduct can be used as a good probe to evaluate the binding of single molecules. The fluorescence quenching of the ct-DNA-EtBr adduct was examined to assess the effect of the sodium sulfonate groups on the binding capabilities of the copper complex in the ct-DNA binding process. The ability of the complexes to quench the EtBr-DNA system in this method is connected to the binding strength of the complexes to the DNA, and gives an indication about the mode of binding (the intercalation process) [42, 43]. The concentration of the EtBr-DNA mixture was maintained at a constant value while the concentration of the complexes was increased by 20 µL aliquots (Fig. 3). The titration process revealed that Cu-L partially quenches the ct-DNA-EtBr adduct while Cu-LSO3Na has no effect on the EtBr-DNA emission. This difference is due to the ability of Cu-L to bind to ct-DNA, displacing some of EtBr units and diminishing the emission intensity. In contrast, the presence of Cu-LSO3Na in the anionic form in the solution causes a repulsive interaction between the copper complex and the negatively charged ribose chain of ct-DNA.
The quenching process does not provide conclusive information on the binding mode. However, the trend in the changes of the viscosity of ct-DNA can indicate the compound-DNA binding mode [42, 43]. Herein, the relative viscosity of ct-DNA has been monitored while increasing the concentrations of Cu-L and Cu-LSO3Na, as well as ethidium bromide as a positive control for benchmarking (Fig. 4). The relative viscosity of the ct-DNA only increased appreciably when increasing the concentration of complex Cu-L, while Cu-LSO3Na showed no pronounced change in the ct-DNA viscosity, which suggests either no interaction at all or groove binding [44]. Taking into consideration that Cu-LSO3Na has no effect on the ct-DNA-EtBr adduct, the complex is likely unable to bind to ct-DNA due to the overall negative charge of the complex that induces a repulsive effect with the negatively charged ribose phosphate chain [45].
3.3 HSA Binding
The interaction of chemotherapeutic agents with blood plasma proteins has been a subject of considerable interest in recent years due to their role in drug transport and metabolism, mainly with serum albumin (about 55% of the blood plasma protein) [46, 47]. Figure 5 illustrates the interaction of complexes with HSA from the concentration dependence of the change in the fluorescence intensity (at 335–340 nm) of the protein upon the addition of complexes at 295 K. The characteristic broad emission band decreases dramatically with small additions of the complexes (Cu-L and Cu-LSO3Na), confirming the interaction between complexes and HSA. The fluorescence quenching is described by the Stern–Volmer equation (Eq. 1); the Stern–Volmer constant can be related to the quenching constant by the following equation:
where kq is the quenching rate constant and τ0 is the average fluorescence lifetime of the protein without the quencher (~ 7 ns) [48, 49]. The data of the two complexes are summarized in Table 1. The quenching constants suggest the existence of the static quenching mechanism as our complexes have rates much higher than 2.00 × 1010 M−1 s−1 (the maximum value for dynamic quenching) [50]. From the Scatchard equation (Eq. 3), the binding constants were calculated, revealing that Cu-L exhibits higher binding constants for HSA than does Cu-LSO3Na (Table 1). The average binding site count for the complexes is nearly 2 for HSA. Indeed, the number of binding sites is more accurately describing the stoichiometric ratio of the compounds to HSA [51]. HSA consists of three domains (DI, DII and DIII), of which two domains are primarily hydrophobic regions which can involve the binding of our complexes [47, 52, 53]. Both compounds showed negative values of ΔG0, indicating the spontaneous interaction with this protein; Cu-L is “more spontaneous” than Cu-LSO3Na (Table 1).
3.4 Molecular Docking
A molecular docking study was undertaken to gain an understanding of the binding sites and possible interactions of the complexes in the protein environment [54, 55]. The lowest ten binding energies of the complexes were obtained, of which the best three are listed in Table 2. In general, Cu-L has a better binding score toward HSA than that calculated for Cu-LSO3Na. The data indicate that Cu-L has a strong preference to bind at site 1 (in DII) (Fig. 6) while Cu-LSO3Na exclusively binds to site 2 (DIII) (Fig. 6). In site 1, the HSA pocket (DII) is surrounded by Glu153, Phe156, Phe157, Arg160, Glu184, Glu188, Ala191, Ser192, Lys195, His 288, Glu292, Glu294, Lys436, His440 and Tyr452 amino acids; some of these amino acid residues are involved in the electrostatic, hydrophobic and van der Waals interactions of Cu-L, with the arginine, in particular, establishing cation-pi interactions. The HSA site 2 pocket (DIII) is on the face of an area consisting of the following amino acids: Asp108, Asn109, Pro110, Asn111, Leu112, Pro113, Arg114, Arg145, Thr422, Glu425 and Leu463; Cu-LSO3Na establishes electrostatic, hydrophobic and van der Waals interactions with some of these amino acids. The dramatic difference in the binding sites with different amino acids for both complexes indicates the crucial nature of the overall net charge of the complexes. Cu-L with its net positive charge is able to squeeze into a pocket with negatively charged amino acids (glutamic acid and aspartic acid) while Cu-LSO3Na forms negatively charged species after dissolving in aqueous medium, and is consequently attracted towards the positively charged arginine amino acid.
3.5 Anticancer Activities
Both complexes were tested against ovcar-3 and hope-62 cell lines. Cu-L has IC50 values of 11.07 ± 0.39 and 21.55 ± 0.69 while Cu-LSO3Na exhibits values of 14.59 ± 0.77 and 16.56 ± 0.57 against ovcar-3 and hope-62, respectively. Comparing the values of both compounds suggests mechanisms involving no DNA binding because Cu-LSO3Na has cytotoxic effects comparable to Cu-L while it has no DNA-binding capability; indeed, the net charge seems to play a minor role in the anticancer activities. However, both complexes have less cytotoxic effect than CuBr(PPh3)(dppz-11-CN) [13] or [Cu(PPh3)2(dppz)]+ [30], which indicates that factors such as steric hindrance and hydrophobicity are more dominant in the anticancer activities of copper(I) complexes.
4 Conclusion
Previous work has suggested that 5,6-diphenyl-3-[2-pyridyl]-1,2,4-triazine (L) ligand enhances the anticancer activities of copper complexes of the formula CuBr(PPh3)(N^N) [12]. In drug design, water-solubility is desirable and hence we sought to evaluate the impact of incorporating the water solubilizing groups sodium sulfonate (5,6-diphenyl-3-[2-pyridyl]-1,2,4-triazine-4,4′-disulfonic acid disodium salt) (L-SO3Na) on the DNA-binding, HSA-binding and anticancer properties. [Cu(PPh3)2(L)]NO3 and [Cu(PPh3)2(L-SO3Na)]NO3 were synthesized and characterized by NMR and IR spectroscopy and mass spectrometry. Absorption and emission spectra of the complexes are almost identical and hence they have minimal difference in their electronic properties. The major difference between the complexes is the net charge: the first complex exists as a cationic species in solution, while the second one possesses a negative-net charge. Ct-DNA binding studies show the strong influence of the net charge as Cu-L is able to bind to ct-DNA while Cu-LSO3Na is not. The net charge of the complexes causes different thermodynamic and kinetic binding capabilities toward human serum albumin. The compounds-HSA binding was further investigated by molecular docking; the two complexes show different preferred binding sites on the HSA protein as a function of their net charge. Finally, the cytotoxicity towards ovcar-3 and hope-62 cancer cell lines reveal that the complexes have slight differences (± 30%) in their cytotoxic effects, indicating that the net charge has some impact on the anticancer activities. However, it seems that DNA is not the major target for this class of complexes. In conclusion, water solubility is desirable, but consideration of the overall net charge of the pharmacophore (anticancer agent) must also be made.
References
E. Wong, C.M. Giandomenico, Current status of platinum-based antitumor drugs. Chem. Rev. 99, 2451–2466 (1999)
C.A. Rabik, M.E. Dolan, Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 33, 9–23 (2007)
P. Heffeter, U. Jungwirth, M. Jakupec, C. Hartinger, M. Galanski, L. Elbling, M. Micksche, B. Kepper, W. Berger, Resistance against novel anticancer metal compounds: differences and similarities. Drug Resist. Updates 11, 1–16 (2008)
N.P. Farrell, Multi-platinum anti-cancer agents. Substitution-inert compounds for tumor selectivity and new targets. Chem. Soc. Rev. 44, 8773–8785 (2015)
S. Medici, M. Peana, V.M. Nurchi, J.I. Lachowicz, G. Crisponi, M.A. Zoroddu, Noble metals in medicine: latest advances. Coord. Chem. Rev. 284, 329–350 (2015)
F. Trudu, F. Amato, P. Vaňhara, T. Pivetta, E.M. Peña-Méndez, J. Havel, Coordination compounds in cancer: past, present and perspectives. J. Appl. Biomed. 13, 79–103 (2015)
T. Lazarevic, A. Rilak, Z.D. Bugarcic, Platinum, palladium, gold and ruthenium complexes as anticancer agents: current clinical uses, cytotoxicity studies and future perspectives. Eur. J. Med. Chem. 142, 8–31 (2017)
C. Marzano, M. Pellei, F. Tisato, C. Santini, Copper complexes as anticancer agents. Anti-Cancer Agents Med. Chem. 9, 185–211 (2009)
I. Iakovidis, I. Delimaris, S.M. Piperakis, Copper and its complexes in medicine: a biochemical approach. Mol Biol. Int (2011). https://doi.org/10.4061/2011/594529
C. Santini, M. Pellei, V. Gandin, M. Porchia, F. Tisato, C. Marzano, Advances in copper complexes as anticancer agents. Chem. Rev. 114, 815–862 (2014)
V. Gandin, M. Porchia, F. Tistao, A. Zanella, E. Severin, A. Dolmella, C. Marzano, Novel mixed-ligand copper(I) complexes: role of diimine ligands on cytotoxicity and genotoxicity. J. Med. Chem. 56, 7416–7430 (2013)
B.A. Babgi, K.H. Mashat, M.H. Abdellattif, M.N. Arshad, K.A. Alzahrani, A.M. Asiri, J. Du, M.G. Humphrey, M.A. Hussien, Synthesis, structure, DNA-binding, cytotoxicity and molecular docking of CuBr(PPh3)(diimine). Polyhedron 192, 114847 (2020)
S. Alsaedi, B.A. Babgi, M.H. Abdellatif, M.N. Arshad, A. Emwas, M. Jaremko, M.G. Humphrey, A.M. Asiri, M.A. Hussein, DNA-binding and cytotoxicity of copper(I) complexes containing functionalized dipyridylphenazine ligands. Pharmaceutics 13, 764–764 (2021)
C. Marzano, M. Pellei, D. Colavito, S. Alidori, G.G. Lobbia, V. Gandin, F. Tisato, C. Santini, Synthesis, Characterization, and in Vitro Antitumor Properties of Tris(hydroxymethyl)phosphine Copper(I) Complexes Containing the New Bis(1,2,4-triazol-1-yl)acetate Ligand. J. Med. Chem. 49, 7317–7324 (2006)
C. Marzano, V. Gandin, M. Pellei, D. Colavito, G. Papini, G.G. Lobbia, E.D. Giudice, M. Porchia, F. Tisato, C. Santini, in vitro antitumor activity of the water soluble copper(I) complexes bearing the tris(hydroxymethyl)phosphine ligand. J. Med. Chem. 51, 798–808 (2008)
M. Porchia, F. Benetollo, F. Refosco, F. Tisato, C. Marzano, V. Gandin, Synthesis and structural characterization of copper(I) complexes bearing N-methyl-1,3,5-triaza-7-phosphaadamantane (mPTA): cytotoxic activity evaluation of a series of water soluble Cu(I) derivatives containing PTA, PTAH and mPTA ligands. J. Inorg. Biochem. 103, 1644–1651 (2009)
M. Porchia, F. Tisato, M. Zancato, V. Gandin, C. Marzano, In vitro antitumor activity of water-soluble copper(I) complexes with diimine and monodentate phosphine ligands. Arab. J. Chem. 13, 998–1010 (2020)
N. Fattahi, A. Ramazani, M. Hamidi, M. Parsa, K. Rostamizadeh, H. Rashidzadeh, Enhancement of brain delivery of methotrexate with administration of mid-chain ester prodrugs: in vitro and in vivo studies. Int. J. Pharmaceutics. 600, 120479 (2021)
N. Fattahi, A. Bahari, A. Ramazani, D. Koolivand, In vitro immunobiological assays of methotrexate-stearic acid conjugate in human PBMCs. Immunobiology 225, 151984 (2020)
N. Fattahi, M.-A. Shahbazi, A. Maleki, M. Hamidi, A. Ramazani, H.A. Santos, Emerging insights on drug delivery by fatty acid mediated synthesis of lipophilic prodrugs as novel nanomedicines. J. Controlled Release 326, 556–598 (2020)
L. Xu, Z. Chu, H. Wang, L. Cai, Z. Tu, H. Liu, C. Zhu, H. Shi, D. Pan, J. Pan, X. Fei, Electrostatically assembled multilayered films of biopolymer enhanced nanocapsules for on-demand drug release. ACS Appl. Bio. Mater. 2, 3429–3438 (2019)
L. Xu, H. Wang, Z. Chu, L. Cai, H. Shi, C. Zhu, D. Pan, J. Pan, X. Fei, X. Lei, Temperature-responsive multilayer films of micelle-based composites for controlled released of a third generation EGFR inhibitor. ACS Appl. Polym. Mater. 2, 741–750 (2020)
Y. Lu, A. Zhuk, L. Xu, X. Liang, E. Kharlampieva, S.A. Sukhishvili, Tunable pH and temperature response of weak polyelectrolyte brushes: role of hydrogen bonding and monomer hydrophobicity. Soft Matter 9, 5464–5472 (2013)
M. Razaghi, A. Ramazani, M. Khoobi, T. Mortezazadeh, E.A. Aksoy, Küçükilinç, Highly fluorinated graphene oxide nanosheets for anticancer linoleic-curcumin conjugate delivery and T2-weighted magnetic resonance imaging: in vitro and in vivo studies. J. Drug Del. Sci. Tech. 60, 101967 (2020)
A. Ebrahimpour, N.R. Alam, P. Abdolmaleki, B.H. Verdom, F. Tirgar, T. Ebrahimi, M. Khoobi, Magnetic metal-organic framework based on zinc and 5-aminolevulinic acid: MR imaging and brain tumor therapy. J. Inorg. Organomet. Polym. Mater. 31, 1208–1216 (2021)
Cotton, F. A.; Goodgame, D. I. L.; Tetrakis(triphenylphosphine)-silver(I) and -copper(I) complexes. J. Chem. Soc. 1960, 5267–5269.
H.T. Al-Masri, A.M. Emwas, Z.A. Al-Talla, M.H. Alkordi, Synthesis and characterization of new N-(diphenylphosphino)naphthylamine chalcogenides: X-ray structures of C10H6-1-HN(Se)Ph2 and Ph2P(S)OP(S)Ph2. Phosphorus Sulfur Silicon Relat. Elem. 187, 1082–1090 (2012)
C.S. Devi, B. Thulasiram, S. Satyanarayana, P. Nagababu, Analytical techniques used to detect DNA binding modes of ruthenium(II) complexes with extended phenanthroline ring. J. Fluoresci. 27, 2119–2130 (2017)
A. Erxleben, Investigation of non-covalent interactions of metal complexes with DNA in cell-free systems. Chimia 71, 102–111 (2017)
W. Villarreal, L. Colina-Vegas, G. Visbal, O. Corona, R.S. Correa, J. Ellena, M.R. Cominetti, A.A. Batista, M. Navarro, Copper(I)-phosphine polypyridyl complexes: synthesis, characterization, DNA/HAS binding study and antiproliferative activity. Inorg. Chem. 56, 3781–3793 (2017)
H. Lineweaver, D. Burk, The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56, 658–666 (1934)
A.S. Abdelhameed, A.H. Bakheit, F.M. Almutairi, H. AlRabiah, A.A. Kadi, Biophysical and in silico studies of the interaction between the anti-viral agents acyclovir and penciclovir, and human serum albumin. Molecules 22, 1906 (2017)
I. Petitpas, A.A. Bhattacharya, S. Twine, M. East, S. Curryi, Crystal structure analysis of warfarin binding to human serum albumin. J. Biol. Chem. 276, 22804–22809 (2001)
N.D. Al-Khathami, K.S. Al-Rashdi, B.A. Babgi, M.A. Hussien, M.N. Arshad, N.E. Eltayeb, S.E. Elsilk, J. Lasri, A.S. Basaleh, M. Al-Jahdali, Spectroscopic and biological properties of platinum complexes derived from 2-pyridyl Schiff basses. J. Saudi Chem. Soc. 23, 903–915 (2019)
N.M. Hosny, M.H. Abdel-Rhman, M.A. Hussien, H.M. Mahmoud, Synthesis, characterization, molecular docking and cytotoxicity studies on N-benzyl-2-isonicotinoylhydrazine-1-carbothioamide and its metal complexes. J. Molec. Struct. 1196, 417–428 (2019)
J.C. Cole, C.W. Murray, J.W. Nissink, R.D. Taylor, R. Taylor, Comparing protein-ligand docking programs is difficult. Proteins 60, 325–332 (2005)
A.N. Jain, Bias, reporting, and sharing: computational evaluations of docking methods. J. Comput. -Aided Mol. Des. 22, 201–212 (2008)
D.N. Muanza, B.W. Kim, K.L. Euler, L. Williams, Antibacterial and antifungal activities of nine medicinal plants of Zaire. Int. J. Pharmacog. 32, 337–345 (1994)
J.M. Pezzuto, C.-T. Che, D.D. McPherson, J.-P. Zhu, G. Topcu, C.A.J. Erdelmeier, G.A. Cordell, DNA as affinity probe useful in the detection and isolation of biologically active natural products. J. Natur. Prod. 54, 1522–1530 (1991)
P. Skehan, R. Storeng, D. Scudiero, A. Monks, J. McMahon, D. Vistica, J. Warren, H. Bokesch, S. Kenney, M.R. Boyd, New colorimetric cytotoxicity as anticancer drug screening. J. Natl. Cancer Inst. 82, 1107–1112 (1990)
A.N. Jadhav, S.B. Pawal, S.S. Chavan, Synthesis, crystal structure and conjugation properties of iminopyridine copper(I) phosphine complex. Inorg. Chim. Acta 440, 77–83 (2016)
Z. Molphy, A. Prisecaru, C. Slator, N. Barron, M. McCann, J. Colleran, D. Chandran, N. Gathergodd, A. Kellett, Copper phenanthrene oxidative chemical nucleases. Inorg. Chem. 53, 5392–5404 (2014)
T. Thirunavukkarasu, H.A. Sparkes, K. Natarajan, Quinoline based Pd(II) complexes: Synthesis, characterization and evaluation of DNA/protein binding, molecular docking and in vitro anticancer activity. Inorg. Chim. Acta 482, 229–239 (2018)
G. Baronea, A. Terenzi, A. Lauriaa, A.M. Almerico, J.M. Leal, N. Bustoc, B. Garcíac, DNA-binding of nickel(II), copper(II) and zinc(II) complexes: Structure–affinity relationships. Coord. Chem. Rev. 257, 2848–2862 (2013)
S.U. Rehman, Z. Yaseen, M.A. Husain, T. Sarwar, H.M. Ishqi, M. Tabish, Interaction of 6-mercaptopurine with calf thymus DNA–deciphering the binding mode and photoinduced DNA damage. PLOS One 9, e93913 (2014)
D. Gibellini, F. Vitone, P. Schiavone, C. Ponti, M.L. Placa, M.C. Re, Quantitative detection of human immunodeficiency virus type 1 (HIV-1) proviral DNA in peripheral blood mononuclear cells by SYBR green real-time PCR technique. J. Clin. Virol. 29, 282 (2004)
J.R. Lakowicz, Principles of Fluorescence Spectroscopy, 3rd edn. (Springer, New York, 2006)
M. Amiri, K. Jankeje, J.R. Albani, Origin of fluorescence lifetimes in human serum albumin. Studies on native and denatured protein. J. Fluor. 20, 651–656 (2010)
M. Amiri, K. Jankeje, J.R. Albani, Characterization of human serum albumin forms with pH. Fluorescence lifetime studies. J. Pharm. Biomed. Anal. 51, 1097–1102 (2010)
W.R. Ware, Oxygen quenching of fluorescence in solution: an experimental study of the diffusion process. J. Phys. Chem. B 66, 455–458 (1962)
E. Lissi, C. Calderon, A. Campos, Evaluation of the number of binding sites in proteins from their intrinsic fluorescence: limitations and pitfalls. Photochem. Photobiol. 89, 1413–1416 (2013)
S. Thangavel, R. Rajamanikandan, H.B. Friedrich, M. Ilanchelian, B. Omondi, Binding interaction, conformational change, and molecular docking study of N-(pyridin-2-ylmethylene)aniline derivatives and carbazole Ru(II) complexes with human serum albumins. Polyhedron 107, 124–135 (2016)
S. Al-harthi, J.I. Lachowicz, M.E. Nowakowski, M. Jaremko, L. Jarmeko, Towards the functional high-resolution coordination chemistry of blood plasma human serum albumin. J. Inorg. Biochem. 198, 110716 (2019)
B. Bhattacharya, S. Nakka, L. Guruprasad, A. Samanta, Interaction of bovine serum albumin with dipolar molecules: fluorescence and molecular docking studies. J. Phys. Chem. B 113, 2143–2150 (2009)
N. Venugopal, G. Krishnamurthy, H.S. Bhojya Naik, J.D. Manohara, DNA binding, molecular docking and antimicrobial evaluation of novel azo dye ligand and their metal complexes. J. Inorg. Organomet. Polym. Mater. 30, 2608–2625 (2020)
Acknowledgements
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, Saudi Arabia, under grant no. (KEP-44-130-40). The authors, therefore, acknowledge with thanks DSR technical and financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alsaedi, S., Babgi, B.A., Abdellatif, M.H. et al. Effect of Net Charge on DNA-Binding, Protein-Binding and Anticancer Properties of Copper(I) Phosphine-Diimine Complexes. J Inorg Organomet Polym 31, 3943–3952 (2021). https://doi.org/10.1007/s10904-021-02063-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10904-021-02063-5