Abstract
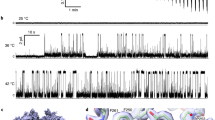
Transient receptor potential vanilloid member 1 (TRPV1) is a Ca2+-permeable cation channel that serves as the primary heat and capsaicin sensor in humans. Using cryo-EM, we have determined the structures of apo and capsaicin-bound full-length rat TRPV1 reconstituted into lipid nanodiscs over a range of temperatures. This has allowed us to visualize the noxious heat-induced opening of TRPV1 in the presence of capsaicin. Notably, noxious heat-dependent TRPV1 opening comprises stepwise conformational transitions. Global conformational changes across multiple subdomains of TRPV1 are followed by the rearrangement of the outer pore, leading to gate opening. Solvent-accessible surface area analyses and functional studies suggest that a subset of residues form an interaction network that is directly involved in heat sensing. Our study provides a glimpse of the molecular principles underlying noxious physical and chemical stimuli sensing by TRPV1, which can be extended to other thermal sensing ion channels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The coordinates are deposited in the Protein Data Bank with PDB IDs 7LP9 (TRPV14C,APO), 7LPA (TRPV14C,CAP), 7LPB (TRPV125C,CAP), 7LPC (TRPV148C,APO), 7LPD (TRPV148C,CAP,INT) and 7LPE (TRPV148C,CAP,OPEN), respectively. The cryo-EM maps are deposited in the Electron Microscopy Data Bank with IDs EMD-23473 (TRPV14C,APO), EMD-23474 (TRPV14C,CAP), EMD-23475 (TRPV125C,CAP), EMD-23476 (TRPV148C,APO), EMD-23477 (TRPV148C,CAP,10sec), EMD-23478 (TRPV148C,CAP,INT) and EMD-23479 (TRPV148C,CAP,OPEN), respectively. Source data are provided with this paper.
References
Caterina, M. J. & Julius, D. The vanilloid receptor: a molecular gateway to the pain pathway. Annu. Rev. Neurosci. 24, 487–517 (2001).
Ramsey, I. S., Delling, M. & Clapham, D. E. An introduction to TRP channels. Annu. Rev. Physiol. 68, 619–647 (2006).
Vriens, J. & Voets, T. Heat sensing involves a TRiPlet of ion channels. Br. J. Pharmacol. 176, 3893–3898 (2019).
Vay, L., Gu, C. & McNaughton, P. A. The thermo-TRP ion channel family: properties and therapeutic implications. Br. J. Pharmacol. 165, 787–801 (2012).
Bandell, M., Macpherson, L. J. & Patapoutian, A. From chills to chilis: mechanisms for thermosensation and chemesthesis via thermo TRPs. Curr. Opin. Neurobiol. 17, 490–497 (2007).
Caterina, M. J. et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824 (1997).
Szallasi, A. et al. Resiniferatoxin-type phorboid vanilloids display capsaicin-like selectivity at native vanilloid receptors on rat DRG neurons and at the cloned vanilloid receptor VR1. Br. J. Pharmacol. 128, 428–434 (1999).
Voets, T. et al. The principle of temperature-dependent gating in cold- and heat-sensitive TRP channels. Nature 430, 748–754 (2004).
Brauchi, S., Orio, P. & Latorre, R. Clues to understanding cold sensation: thermodynamics and electrophysiological analysis of the cold receptor TRPM8. Proc. Natl Acad. Sci. USA 101, 15494–15499 (2004).
Yin, Y. et al. Structure of the cold- and menthol-sensing ion channel TRPM8. Science 359, 237–241 (2018).
Yang, S. et al. A paradigm of thermal adaptation in penguins and elephants by tuning cold activation in TRPM8. Proc. Natl Acad. Sci. USA 117, 8633–8638 (2020).
Paricio-Montesinos, R. et al. The sensory coding of warm perception. Neuron 106, 830–841 (2020).
Yao, J., Liu, B. & Qin, F. Kinetic and energetic analysis of thermally activated TRPV1 channels. Biophys. J. 99, 1743–1753 (2010).
Grandl, J. et al. Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat. Neurosci. 13, 708–714 (2010).
Kim, S. E., Patapoutian, A. & Grandl, J. Single residues in the outer pore of TRPV1 and TRPV3 have temperature-dependent conformations. PLoS ONE 8, e59593 (2013).
Yao, J., Liu, B. & Qin, F. Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc. Natl Acad. Sci. USA 108, 11109–11114 (2011).
Jara-Oseguera, A., Bae, C. & Swartz, K. J. An external sodium ion binding site controls allosteric gating in TRPV1 channels. Elife 5, e13356 (2016).
Zhang, F. et al. Heat activation is intrinsic to the pore domain of TRPV1. Proc. Natl Acad. Sci. USA 115, E317–E324 (2018).
Vlachova, V. et al. Functional role of C-terminal cytoplasmic tail of rat vanilloid receptor 1. J. Neurosci. 23, 1340–1350 (2003).
Yang, F., Cui, Y., Wang, K. & Zheng, J. Thermosensitive TRP channel pore turret is part of the temperature activation pathway. Proc. Natl Acad. Sci. USA 107, 7083–7088 (2010).
Kim, M. et al. Evidence that the TRPV1 S1–S4 membrane domain contributes to thermosensing. Nat. Commun. 11, 4169 (2020).
Clapham, D. E. & Miller, C. A thermodynamic framework for understanding temperature sensing by transient receptor potential (TRP) channels. Proc. Natl Acad. Sci. USA 108, 19492–19497 (2011).
Chowdhury, S., Jarecki, B. W. & Chanda, B. A molecular framework for temperature-dependent gating of ion channels. Cell 158, 1148–1158 (2014).
Jordt, S. E. & Julius, D. Molecular basis for species-specific sensitivity to ‘hot’ chili peppers. Cell 108, 421–430 (2002).
Cao, E., Liao, M., Cheng, Y. & Julius, D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013).
Gao, Y., Cao, E., Julius, D. & Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016).
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013).
Yang, F. et al. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol. 11, 518–524 (2015).
Elokely, K. et al. Understanding TRPV1 activation by ligands: insights from the binding modes of capsaicin and resiniferatoxin. Proc. Natl Acad. Sci. USA 113, E137–E145 (2016).
Darre, L. & Domene, C. Binding of capsaicin to the TRPV1 ion channel. Mol. Pharm. 12, 4454–4465 (2015).
Hanson, S. M., Newstead, S., Swartz, K. J. & Sansom, M. S. P. Capsaicin interaction with TRPV1 channels in a lipid bilayer: molecular dynamics simulation. Biophys. J. 108, 1425–1434 (2015).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Nadezhdin, K. D. et al. Extracellular cap domain is an essential component of the TRPV1 gating mechanism. Nat. Commun. 12, 2154 (2021).
Jordt, S. E., Tominaga, M. & Julius, D. Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc. Natl Acad. Sci. USA 97, 8134–8139 (2000).
Zubcevic, L., Borschel, W. F., Hsu, A. L., Borgnia, M. J. & Lee, S. Y. Regulatory switch at the cytoplasmic interface controls TRPV channel gating. Elife 8, e47746 (2019).
Singh, A. K. et al. Structural basis of temperature sensation by the TRP channel TRPV3. Nat. Struct. Mol. Biol. 26, 994–998 (2019).
Swain, J. & Kumar Mishra, A. Location, partitioning behavior and interaction of capsaicin with lipid bilayer membrane: study using its intrinsic fluorescence. J. Phys. Chem. B 119, 12086–12093 (2015).
Sánchez-Moreno, A. et al. Irreversible temperature gating in trpv1 sheds light on channel activation. Elife 7, e36372 (2018).
Ladrón-de-Guevara, E. et al. The contribution of the ankyrin repeat domain of TRPV1 as a thermal module. Biophys. J. 118, 836–845 (2020).
Ryu, S., Liu, B., Yao, J., Fu, Q. & Qin, F. Uncoupling proton activation of vanilloid receptor TRPV1. J. Neurosci. 27, 12797–12807 (2007).
Bae, C. et al. Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin. Elife 5, e11273 (2016).
Salazar, H. et al. Structural determinants of gating in the TRPV1 channel. Nat. Struct. Mol. Biol. 16, 704–710 (2009).
Steinberg, X. et al. Conformational dynamics in TRPV1 channels reported by an encoded coumarin amino acid. Elife 6, e28626 (2017).
Zubcevic, L. & Lee, S. Y. The role of π-helices in TRP channel gating. Curr. Opin. Struct. Biol. 58, 314–323 (2019).
Susankova, K., Ettrich, R., Vyklicky, L., Teisinger, J. & Vlachova, V. Contribution of the putative inner-pore region to the gating of the transient receptor potential vanilloid subtype 1 channel (TRPV1). J. Neurosci. 27, 7578–7585 (2007).
Makhatadze, G. I. & Privalov, P. L. Energetics of protein structure. Adv. Protein Chem. 47, 307–425 (1995).
Henriques, D. A., Ladbury, J. E. & Jackson, R. M. Comparison of binding energies of SrcSH2-phosphotyrosyl peptides with structure-based prediction using surface area based empirical parameterization. Protein Sci. 9, 1975–1985 (2000).
Franzese, G. & Rubi, M. (eds) Aspects of Physical Biology: Biological Water, Protein Solutions, Transport and Replication (Springer, 2008).
Voets, T. Quantifying and modeling the temperature-dependent gating of TRP channels. Rev. Physiol. Biochem. Pharmacol. 162, 91–119 (2012).
Chen, H., Deng, J., Cui, Q., Chanda, B. & Henzler-Wildman, K. Mapping temperature-dependent conformational change in the voltage-sensing domain of an engineered heat-activated K+ channel. Proc. Natl Acad. Sci. USA 118, e2017280118 (2021).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014).
Ritchie, T. K. et al. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Delano, W. L. The PyMol Molecular Graphics System (DeLano Scientific, 2002).
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007).
Kang, K. et al. Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature 481, 76–80 (2011).
Fraczkiewicz, R. & Braun, W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 19, 319–333 (1998).
Acknowledgements
Cryo-EM data were collected at the Duke University Shared Materials Instrumentation Facility (SMIF) and at the Pacific Northwest Center for Cryo-EM (PNCC) at OHSU. We thank J. Myers at PNCC for assistance with data collection, N. Bhattacharya at SMIF for assistance with the microscope operation, L.B. Dillard at NIEHS for assistance with sample screening, and A. Bartesaghi at Duke for a pre-processing interface for data collection. We thank J. Fedor and Y. Yin for critical reading of the manuscript and discussions, W. Borschel for training F. Zhang on the patch-clamp recording, J. Grandl for discussions and W. Im for advice on the solvent accessibility analysis. This research was supported by NIH grant R35NS097241 (to S.-Y.L.) and by the National Institute of Health Intramural Research Program, US National Institutes of Environmental Health Sciences (ZIC ES103326 to M.J.B). A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. DUKE SMIF is affiliated with the North Carolina Research Triangle Nanotechnology Network, which is in part supported by the NSF (ECCS-2025064).
Author information
Authors and Affiliations
Contributions
D.H.K. conducted biochemical preparation, sample freezing and single-particle 3D reconstruction under the guidance of S.-Y.L. F.Z. carried out all electrophysiological recordings under the guidance of S.-Y.L. Y.S. collected cryo-EM data and helped D.H.K. with cryo-EM data processing. D.H.K. and S.-Y.L. performed model building. J.B. helped D.H.K. in part of the sample screening under the guidance of M.J.B. S.-Y.L. and D.H.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks Chia-Hsueh Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Florian Ullrich was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 TRPV1 data collection and processing.
Data processing procedures, a, Data processing flow chart for TRPV14C,APO, TRPV14C,CAP, TRPV125C,CAP, TRPV148C,APO. b, representative micrographs, see Table 1 for details. c, 2D classification images, d, 3D reconstructions, e, local resolution estimation, f, the Euler distribution plot, g, FSC curves for TRPV14C,APO, TRPV14C,CAP, TRPV125C,CAP, TRPV148C,APO, TRPV148C,CAP,OPEN and TRPV148C,CAP,INT, respectively. h, Data processing flow chart for TRPV148C,CAP,INT and TRPV148C,CAP,OPEN.
Extended Data Fig. 2 Representative Cryo-EM density of the TRPV1 structures.
a-f, cryo-EM density for subdomains in TRPV14C,APO (a, thresholding 0.014), TRPV14C,CAP (b, thresholding 0.014), TRPV125C,CAP (c, thresholding 0.025), TRPV148C,APO (d, thresholding 0.019), TRPV148C,CAP,INT (e, thresholding 0.28), TRPV148C,CAP,OPEN (f, thresholding 0.3). Structural elements are shown as sticks and EM density as gray mesh.
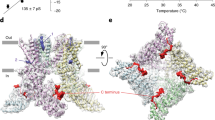
Extended Data Fig. 3 Structural features of the full-length TRPV1.
a, Architecture of the TRPV1 protomer with subdomains indicated: ankyrin repeat domain (ARD), coupling domain (CD), transmembrane helices S1-S6, TRP helix, and C-terminal domain (CTD). b, Cryo-EM density (half-map without symmetry) for the selectivity filter of TRPV14C,APO corresponding to putative sodium ions at 0.04 thresholding. c, Cryo-EM density of the turret and turret junction (0.012 thresholding). d, Close-up view of the outer pore and turret junction (0.012 thresholding). e, Interaction networks spanning the outer pore region and the S1-S4 domain (0.03 thresholding). Key residues interacting with E600 and E648 are shown as sticks with surrounding cryo-EM density. f, g, Cryo-EM density of the CD, TRP, ARD (f), and CTD (g). The ARD is colored in gold, the CD and its individual elements (HTHCD, βCD,) in sky blue, the TRP domain in dark green, and the CTD in orange. The cryo-EM density (gray) is shown at 0.012 thresholding. h, Superposition of a single protomer from TRPV14C,APO (blue) and TRPV148C,APO (gold). i, Superposition of a single protomer from TRPV14C,APO (blue) and TRPV14C,CAP (cyan).
Extended Data Fig. 4 Comparison of TRPV14C,APO and TRPV148C,APO.
a, b, Cryo-EM 3D reconstructions of TRPV14C,APO (a, blue) and TRPV148C,APO (b, gold), respectively. Outlines indicate AR1-AR4. c, Close-up comparison of the cytoplasmic domains between TRPV14C,APO (blue) and TRPV148C,APO (gold).
Extended Data Fig. 5 Comparison of TRPV14C,APO, TRPV14C,CAP, TRPV125C,CAP and the published structure of TRPV1 in the presence of capsaicin.
a, Close-up view of the S1-S4 domain of TRPV14C,APO (blue) and TRPV14C,CAP (cyan). Capsaicin (red) and phosphatidyl inositol (blue) molecules are shown as sticks. b, Close-up view of capsaicin in the vanilloid pocket of TRPV14C,CAP. The cryo-EM density is shown at 0.025 thresholding. c, Side view comparison of TRPV14C,CAP (cyan) and TRPV125C,CAP (green). d, Side view comparison of TRPV14C,CAP (green) and the published TRPV1 structure in the presence of capsaicin (PDB ID: 3J5R, brown).
Extended Data Fig. 6 Comparison between the overall structures of TRPV14C,APO, TRPV148C,CAP,OPEN and DkTx/RTx-bound TRPV1.
a, Comparison of TRPV14C,APO (silver), TRPV148C,CAP,OPEN (red), and DkTx/RTx-TRPV1 (blue) viewed from the intracellular side. ARD/CD movement occurs at an individual protomer level. b, Comparison of the S6b and TRP domain of TRPV14C,APO, TRPV148C,CAP,OPEN, and DkTx/RTx-TRPV1. c, Close-up view of TRPV14C,APO, TRPV148C,CAP,OPEN, and DkTx/RTx-TRPV1 in the cytoplasmic domains. d, Alternate angle and close-up view of TRPV14C,APO, TRPV148C,CAP,OPEN, and DkTx/RTx-TRPV1 in the cytoplasmic domains.
Extended Data Fig. 7 Comparison of TRPV148C,CAP,OPEN and DkTx/RTx-bound TRPV1 structures.
a, The overlapping locations of phospholipid (TRPV148C,CAP,OPEN, red) and DkTx (DkTx/RTx-TRPV1, blue), shown as sticks and spheres, between the pore loop and pore helix. Several side chains are shown as sticks to illustrate the differences in the outer pore of the two structures. b, Structural differences between TRPV148C,CAP,OPEN and DkTx/RTx-TRPV1 at S6, the S4-S5 linker, and the TRP helix.
Extended Data Fig. 8 Solvent accessible surface area-based heat capacity change plots for the first and the second transitions.
a, b, ΔCPpred plots for the first (a) and second (b) transitions. For each transition, residues exhibiting positive ΔCPpred are plotted in the upper graph using log10(ΔCPpred), and residues exhibiting negative ΔCPpred are plotted in the lower graph using -log10(-ΔCPpred). The dotted line denotes the 15 J mol-1 K-1 threshold. ΔCPpred was calculated as described in the Methods. Residues for which the side chains were not resolved were not included in the calculation.
Extended Data Fig. 9 Rearrangement in the vanilloid pocket during the heat-dependent transitions.
a, Close-up view of the vanilloid binding site in TRPV125C,CAP (green), TRPV148C,CAP,INT (orange), and TRPV148C,CAP,OPEN (red). Several key residues in capsaicin are shown as sticks. Dotted lines denote either H-bond or salt bridge interactions. The 310 helical region of S4 is indicated as 310. b, Close-up view of S5 and S6 in TRPV148C,CAP,INT (orange), and TRPV148C,CAP,OPEN (red). The π helical turn in S6 is denoted by π. c, Comparison of TRPV148C,CAP,OPEN (red) and DkTx/RTx-bound TRPV1 (PDB ID: 5IRX, blue). DkTx is shown as sticks and gray spheres; capsaicin is depicted as sticks only.
Supplementary information
Source data
Source Data Fig. 2
Statistical source data for the electrophysiology experiment.
Source Data Fig. 5
Statistical source data for the electrophysiology experiment.
Source Data Fig. 6
Statistical source data for the electrophysiology experiment.
Rights and permissions
About this article
Cite this article
Kwon, D.H., Zhang, F., Suo, Y. et al. Heat-dependent opening of TRPV1 in the presence of capsaicin. Nat Struct Mol Biol 28, 554–563 (2021). https://doi.org/10.1038/s41594-021-00616-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00616-3
This article is cited by
-
Looking back at 30 years of Nature Structural & Molecular Biology
Nature Structural & Molecular Biology (2024)
-
Research focus and thematic trends of transient receptor potential vanilloid member 1 research: a bibliometric analysis of the global publications (1990–2023)
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Relation between flexibility and intrinsically disorder regions in thermosensitive TRP channels reveal allosteric effects
European Biophysics Journal (2024)
-
Transient Receptor Potential Channels: Multiple Modulators of Peripheral Neuropathic Pain in Several Rodent Models
Neurochemical Research (2024)
-
Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands
Nature Chemical Biology (2023)