Corals and Sponges Under the Light of the Holobiont Concept: How Microbiomes Underpin Our Understanding of Marine Ecosystems
- InBioS – Animal Physiology and Ecophysiology, Department of Biology, Ecology & Evolution, University of Liège, Liège, Belgium
In the past 20 years, a new concept has slowly emerged and expanded to various domains of marine biology research: the holobiont. A holobiont describes the consortium formed by a eukaryotic host and its associated microorganisms including bacteria, archaea, protists, microalgae, fungi, and viruses. From coral reefs to the deep-sea, symbiotic relationships and host–microbiome interactions are omnipresent and central to the health of marine ecosystems. Studying marine organisms under the light of the holobiont is a new paradigm that impacts many aspects of marine sciences. This approach is an innovative way of understanding the complex functioning of marine organisms, their evolution, their ecological roles within their ecosystems, and their adaptation to face environmental changes. This review offers a broad insight into key concepts of holobiont studies and into the current knowledge of marine model holobionts. Firstly, the history of the holobiont concept and the expansion of its use from evolutionary sciences to other fields of marine biology will be discussed. Then, the ecology and physiology of marine holobionts will be investigated through the examples of corals and sponges. We will discuss the impacts of environmental change on organisms at the holobiont level and how microbiomes contribute to the resilience and/or vulnerability of their host in the face of environmental stressors. Finally, we will conclude with the development of new technologies, holistic approaches, and future prospects for conservation biology surrounding marine holobionts.
The Marine Holobiont Concept
A Short History of a New Concept
The first time the term “symbiosis” (see Box 1 for full definition of this term and others used in this review article) was used in a biological context dates from 1878 when the German botanist and mycologist, Heinrich Anton de Bary, defined the symbiosis as “the living together of differently named organisms” in its lecture entitled “Die Erscheinung der Symbiose” (“The phenomenon of symbiosis”) (De Bary, 1879; Oulhen et al., 2016). In the following decades, the idea that different organisms establish partnerships and live in association slowly implemented itself in scientific literature. In the mid-20th century, theoretical biologist Adolf Meyer-Abich developed the “theory of holobiosis,” notably arguing that it was necessary to focus on describing the processes of assemblage of independent organisms to explain evolutionary change (Baedke et al., 2020). This evolutionary theory was the precursor to the modern notion of “holobiont” introduced in Margulis and Fester (1991). Deriving the concept from their work on endosymbiosis, they described a holobiont as “a simple biological entity involving a host and a single inherited symbiont” (Margulis and Fester, 1991; Simon et al., 2019). Today, the notion of holobiont has been extended to define a host and its associated communities of microorganisms, and the concept is widely used in various biological research fields, from invertebrate and plant holobionts to humans (Vandenkoornhuyse et al., 2015; Kundu et al., 2017; Simon et al., 2019). A quick search through scientific publication databases shows how popular the term “holobiont” has become in the past 10 years, with a steady exponential increase since the beginning of the 2000s (Figure 1). This marked rise in holobiont studies goes in pair with the blossoming of high throughput sequencing (HTS) technologies during the past 20 years. Indeed, since the completion of the human genome project in 2003, sequencing technologies have experienced extraordinary progress with the introduction of next generation sequencing (NGS), leading to a significantly decreased cost per megabase and reduced analysis time. Today, NGS technologies have become a routine part of biological research (Goodwin et al., 2016). Microbiome studies investigating the composition, functions and dynamics of complex microbial communities inhabiting diverse environments have multiplied, and with this, microbiome analysis methods and standards have rapidly advanced (Knight et al., 2018). The ocean microbiome has been widely studied for several decades and the emergence of holistic -omics approaches became central to this research field to establish the link between microbial diversity and microbial functions in the ocean (Moran, 2015; Coutinho et al., 2018). Similarly, HTS and -omics methodologies are increasingly employed to study the structure and functions of complex associations between microbial communities and their marine host. Between 2016 and 2018, the Tara Pacific expedition collected metadata and applied state-of-the-art technologies in very high throughput genetic sequencing and molecular analysis to uncover the microbial, chemical and functional diversity associated with coral holobionts (Planes et al., 2019). Recent advances in biological technologies have allowed the development of a novel strategy in the study of marine ecosystems and organisms: the holobiont approach. Since the first idea of symbiosis and theories of holobiosis in the 19th and 20th centuries, we finally have the means to investigate these complex relationships and work in a novel framework.
BOX 1. Glossary.
Adaptation sensu lato: Describes any transgenerational process that increases the fitness of the holobiont phenotype. It includes: physiological transgenerational acclimation, host evolution (often involving epigenetic changes), heritable microbial community changes, and microbial evolution (Webster and Reusch, 2017).
Adaptation sensu stricto: The equivalent of evolutionary adaptation. Describes the hereditary alteration or adjustment in the mean phenotype of a population due to natural selection and involving genetic change in the form of differing allele frequencies between generations.
Assisted evolution: Describes the range of practices involving active human intervention to accelerate the rate of naturally occurring evolutionary processes. The aim is to enhance certain attributes to help species adapt to changing environmental conditions faster than they would via natural selection.
Beneficial microorganisms: Microorganisms that improve the fitness of others, and, in the context of holobionts, of the host they live in association with.
Codiversification: Simultaneous diversification (evolution) of two species lineages.
Coevolution: Reciprocal evolutionary change between interacting species that exert selection pressures on each other.
Coral probiotic hypothesis: Hypothesis stating that a dynamic relationship exists between symbiotic microorganisms and environmental conditions which prompts the selection of the most advantageous coral holobiont. Corals adapt to changing environmental conditions more rapidly (days to weeks) by changing their microbial partners than through mutation and selections (years) (Reshef et al., 2006).
Core microbiome: Members of a host’s microbial community that are highly prevalent across all individuals of the same species and whose functions are potentially vital for holobiont functioning.
Disease: Any impairment to cells or tissues of an organism that results in its dysfunction.
Dysbiosis: Changes in microbiome composition frequently correlated with disease states, without inferring a causative link between the two.
Ecogenomics: The study of the ecology of microorganisms through the analysis of genomes and metagenomes in the aim to understand ecosystem functioning through the microbial lens.
Ecosystem engineers: Organisms that create, maintain and/or modify habitats with impacts on the availability of resources to other species by being the drivers of physical and chemical changes to the environment. Corals and sponges are excellent examples of marine ecosystem engineers in environments such as coral reefs or deep-sea sponge grounds.
Endosymbionts: Organisms that entertain a symbiotic relationship within a host, either inside the cells (endocytosymbionts, intracellular symbionts, or endocellular symbionts) or attached to the surface of cells (extracellular endosymbionts).
Holobiont: Eukaryotic host with its endocellular and extracellular microbiome including bacteria, archaea, protists, fungi, and viruses.
Hologenome: The sum of the genetic information of the host and its associated symbiotic microorganisms.
Hologenome theory of evolution: Theory of evolution that considers the holobiont (animal or plant with all of its associated microorganisms) as a unit of selection in evolution.
Horizontal gene transfer (HGT): The transfer of genetic material between non-mating species, often a process leading to genetic differentiation, adaptive evolution, and favoring coevolution.
Horizontal transmission: Acquisition of microorganisms (symbiotic, commensal or parasitic) from the surrounding environment or from a nearby host.
Microbiome: Community of microorganisms found at a specific place and/or a specific time.
Metaorganism: Eukaryotic host and associated microorganisms for which the function is known or implied at a moment in time and in a given environment.
Nested ecosystem: A biological community of interacting organisms and their physical environment nested within a larger one. Each ecosystem is an integrated whole as well as a part of larger systems. Changes within an ecosystem can affect the properties of the ecosystems nested within it and the larger ecosystems in which it exists.
Phylosymbiosis: The retention of the phylogenetic signal of a host within its associated microbial community through the correlation between host phylogenetic relatedness and multivariate community similarities of the associated microbiome.
Marine probiotics: Marine bacterial strains that favor the growth of beneficial microbes within microbiomes and that restrict the proliferation of decay- or disease-causing pathogens.
Sponge loop: Recycling of the dissolved organic matter (DOM) produced by corals and algae into the food web through sponge activity.
Symbiosis: Persistent and intimate relationship between two or more dissimilar organisms from which at least one of them benefits.
Variable microbiome: Members of a host’s microbial community that are present only in some individuals of a same species or that vary in their relative abundance.
Vertical transmission: Direct transmission of microorganisms (symbiotic, commensal, or parasitic) from parent to offspring.
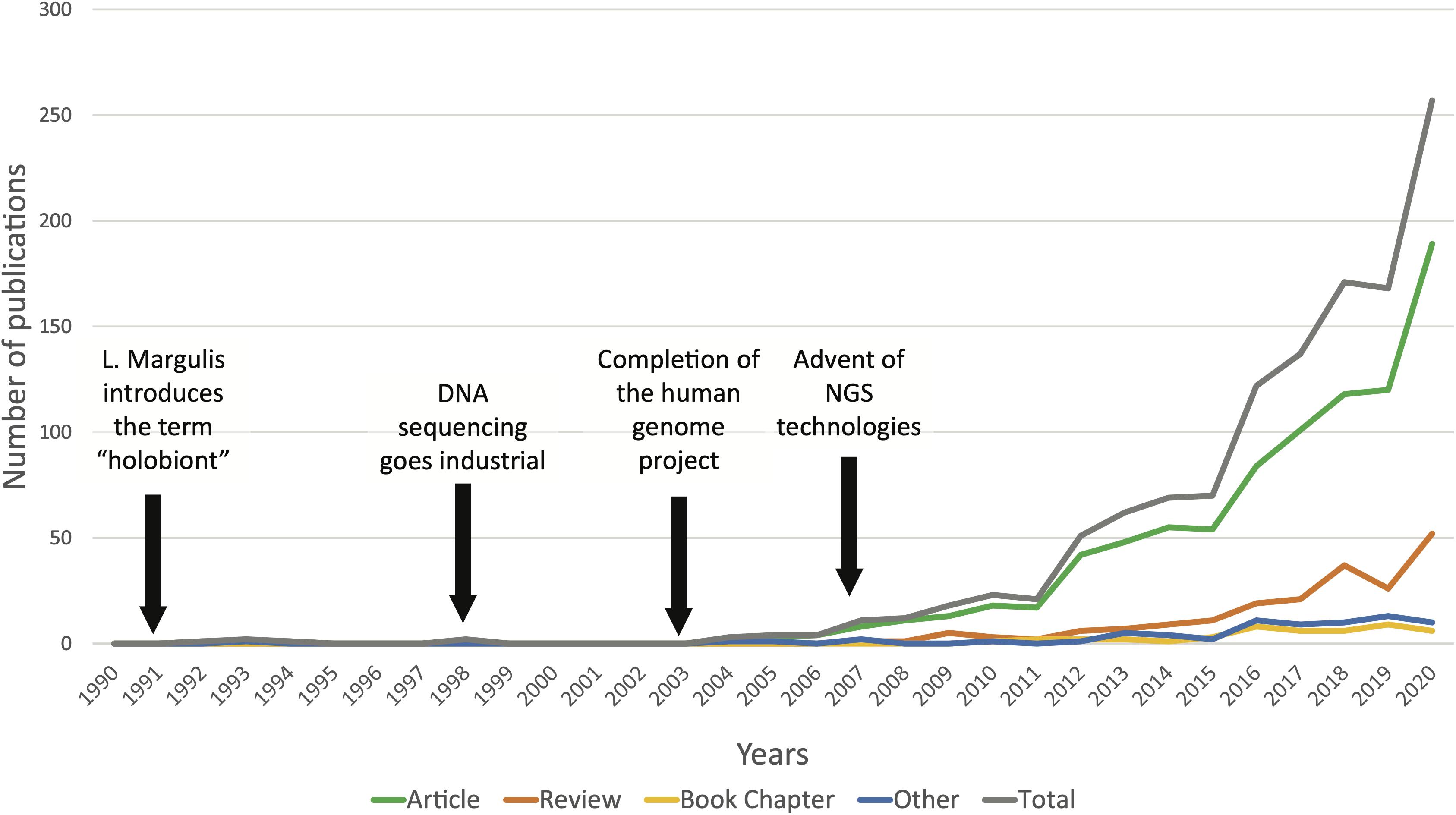
Figure 1. Occurrence of the term “holobiont” in the title, abstract, and/or keywords of scientific publications from 1990 to December 2020 (N = 1209). Data retrieved from the curated citation and abstract database Scopus.
Key Concepts in Holobiont Studies
The holobiont concept emerged from an evolutionary framework but it was progressively used in different contexts such as physiological and host–microbiome interactions, ecological and biotic interactions in complex ecosystems. Some of the key concepts in global holobiont studies emerged from the study of coral holobionts. In 2006, the observation that corals could adapt to environmental stressors such as elevated temperatures and infections by specific pathogens led to the emergence of the coral probiotic hypothesis. This hypothesis states that the dynamic relationship existing between symbiotic microorganisms and environmental conditions leads to the selection of the most advantageous coral holobiont (Reshef et al., 2006). Indeed, the ability of corals to modulate their microbial partners allows them to adapt more promptly to changing environmental conditions than by mutation and selection.
Extrapolating from the coral probiotic hypothesis, Ilana Zilber-Rosenberg and Eugene Rosenberg introduced the concept of “hologenome” for all invertebrates, higher animals and plants, and proposed a higher order of postulation: the hologenome theory of evolution (Rosenberg et al., 2007; Zilber-Rosenberg and Rosenberg, 2008). The hologenome is simply defined as the sum of the genetic information of the host and its associated symbiotic microorganisms. The hologenome theory of evolution posits that the holobiont with its hologenome, acting in consortium, should be considered as the unit of natural selection in evolution, and that the microbial symbionts have an important role in the adaptation and evolution of the holobiont through their relatively rapid variations in diversity (Rosenberg et al., 2007; Zilber-Rosenberg and Rosenberg, 2008). However, the hologenome concept needs to be used with care. Some opponents to this theory argue that the hologenome is based on overly restrictive assumptions as it requires high partner fidelity for the entire host–microbiome if it is to evolve as a unit; and where it does not, there is the potential for complex interactions and effects leading to both mutualistic and antagonistic (fitness conflict) evolution (Douglas and Werren, 2016). Others warn against interpreting evolution solely through the lens of the hologenome as intimate and highly specific relationships between host and microbiome do not necessarily entail coevolution (Moran and Sloan, 2015). Indeed, it is vital to distinguish between the concepts of coevolution, codiversification, and phylosymbiosis when talking about evolutionary processes at the holobiont level. Coevolution occurs when interacting species affect each other’s evolution through the process of natural selection. This can occur between two species (pairwise coevolution), a suite of species (diffuse coevolution) and can involve gene-for-gene correspondence among species (matching gene coevolution) (Langerhans, 2008). Codiversification is the simultaneous diversification (evolution) of two species lineages. However, codiversification does not necessarily imply coevolution (Moran and Sloan, 2015). Phylosymbiosis is a fairly recent concept introduced to designate the phenomenon of microbial community relationships that recapitulate the phylogeny of their host (Brucker and Bordenstein, 2013; Lim and Bordenstein, 2020). It is a pattern observed at one moment in time and space and does not assume a stable evolutionary association or congruent ancestral splits between a host and its associated microbiome (O’Brien et al., 2020).
Ten years after the original proposal of the hologenome theory of evolution, the authors refined their position concerning the principles of evolution and the available data supporting them. They conclude that considerable evidence exists to support the hypothesis that the holobiont, with its hologenome, is a level of selection in evolution as it interacts, replicates, manifests adaptation and, benefits from the selection process (Rosenberg and Zilber-Rosenberg, 2018; Roughgarden et al., 2018). Rapid genomic changes in the microbiome allow holobionts to constantly adapt to changing environmental conditions. This would provide the time necessary for the host genome to adapt and evolve (Rosenberg and Zilber-Rosenberg, 2018). Holobionts and hologenomes are undeniable multipartite entities resulting from ecological, evolutionary and genetic processes at different levels; they do not only refer to a simple process but constitute a wider structure for host biology in the light of the microbiome (Theis et al., 2016). The concept of the holobiont constitutes an important reminder that there is a reciprocal impact of each partners’ growth and survival on the evolution of both the host and associated microbial symbionts. However, attention must be kept to the behavior of individual species that comprise communities while considering the significance of their interactions for the ecology and evolution of both the host and microbial partners (Koskella and Bergelson, 2020). More importantly, working within the holobiont-hologenome framework is acknowledging the innate complexity of biological systems and adopting a holistic approach to understand the evolution, physiology, and ecology of organisms.
Holobionts in Marine Systems and Nested Ecosystems
Microbiome research has revolutionized the way scientists reflect on the roles of microorganisms in ecosystem functions in a variety of environments, from plants in terrestrial ecosystems (Vandenkoornhuyse et al., 2015), to animal and human guts (Kundu et al., 2017), to the diverse marine ecosystems. Holobiont research in the marine environment is particularly pertinent since microorganisms dominate the ocean in biomass, diversity and metabolic activity. They are major drivers in biogeochemical cycles, they colonize every ecosystems and living organisms, thus considerably influencing their functioning and health (Arrigo, 2005; DeLong, 2009; Moran, 2015). The physical nature of water allows for greater physicochemical connectivity between habitats and the organisms they host, resulting in flexible ecosystems where dispersal barriers are reduced and microbial diversity shifts are facilitated compared to terrestrial environments (Kinlan and Gaines, 2003; Dittami et al., 2020).
In addition to the roles marine microorganisms fulfill in geochemical and nutrient cycling at an ecosystem-wide scale, they support the health of a wide array of benthic and pelagic organisms at a cellular and molecular scale through complex associations and symbiotic interactions (Rosenberg et al., 2007; Webster and Taylor, 2012; Egan et al., 2013; Leitão et al., 2020; Vanwonterghem and Webster, 2020). The recent uncovering of the astounding marine microbial diversity is moving marine biology research into the study of the interactions between the host and a multi-member microbiome. Holobiont research in the marine environment involves multiple levels of investigation, from understanding the intricate mechanisms of communication and cycling of nutrients between species from different kingdoms, predicting the consequences for the health of the holobiont, to the repercussions at the ecosystem levels. Holobionts are complex units within which the actions and interactions of their components affect the overall state of the holobiont. In this way, holobionts can be considered as functioning ecosystems that are nested within successively larger systems where they interact at a wider scale and influence neighboring holobionts (Pita et al., 2018; Vanwonterghem and Webster, 2020). Pita et al. (2018), in their review on the sponge holobiont, introduced the concept of nested ecosystem which was further exploited by Vanwonterghem and Webster (2020) in the context of coral reefs communities. This concept implies that the microbiome functions at the organismal level modulate holobiont performance which, in turn, influences its interactions with the surrounding environment. In this way, functional constituents of the microbiome impact ecosystem health and functioning through cascading effects involving primary production, nutrient cycling, disease regulation, and community structure (Pita et al., 2018; Vanwonterghem and Webster, 2020). In a similar train of thoughts, the concept of “eco-holobiont” was recently proposed to describe the system of interactions between biotic (plants and animals) and environmental (e.g., water, soil, sediment, air) microbiomes which constitute a microbial loop that modulates the functioning of an assembly of holobionts within an ecosystem (Singh et al., 2020). Some advocate for the use of the term “metaorganism” to describe the holobiont and the associated microorganisms whose functions are recognized at a moment in time and in a specific environment. In contrast, the term holobiont is then simply used to describe the host and its entire associated microbial diversity, while keeping in mind that not all microorganisms are functionally important and that associated microorganisms are not involved in all host processes (Jaspers et al., 2019).
Studying holobionts in marine ecosystems is inherently complex due to the multilevel nature of the interactions between organisms from different kingdoms, in a spatiotemporal context that is highly sensitive to environmental perturbations. However, adopting the holobiont concept in marine biological research is of the utmost importance. Indeed, some of the most important ecosystem engineers such as reef-building corals, sponges, deep-sea mussels or hydrothermal vent tubeworms rely heavily on their microbial partners to sustain primary productivity, nutrient flows and create the structural habitats and resources supporting their respective ecosystem (Wilkins et al., 2019). Holobiont research is particularly gaining popularity in coastal marine environments that provide numerous ecosystem services. Trevathan-Tackett et al. (2019) identified key marine microbiome research themes to be urgently addressed to gain a deeper understanding of coastal ecosystem dynamics and functions framed under the holobiont concept. The themes include linking microbiome dynamics with spatiotemporal parameters, linking community structure with microbial function and the impacts on the resilience and health of the holobiont, understanding host-microbiome reciprocal interactions and tripartite interaction with the environment, and reflecting on how we can manage microbiomes in the coastal environment and the ways marine holobionts are relevant to human health and well-being (Trevathan-Tackett et al., 2019).
Holobiont Models in Marine Environments
To best characterize “functional holobionts,” otherwise referred to as metaorganisms, some authors prone the study of a limited set of model organisms for which clear methodologies can be established and whose associated microbes can be manipulated in a less complex environment (Jaspers et al., 2019). Such model organisms include early diverging non-bilaterian metazoans such as Hydra (freshwater model), Nematostella, and Exaiptasia (Jaspers et al., 2019) or other organisms that can be cultivated ex situ aposymbiotically for fully controlled experiments such as the upside-down jellyfish Cassiopea, the bobtail squid Euprymna scolopes, the flatworm Symsagittifera roscoffensis, the unicellular alga Ostreococcus or the green macroalga Ulva (Dittami et al., 2020). The freshwater Hydra model forms a tripartite partnership with symbiotic algae, Chlorella, and stably associated microorganisms which can be studied in a controlled environment to decipher the nature of the interactions, the interkingdom communication tools such as the Toll-Like Receptors (TLR) or the dynamics of the holobiont following environmental perturbations (Bosch, 2012; Bathia and Bosch, 2020). The renowned Vibrio-squid model has allowed to closely investigate bacterial-animal interactions from the establishment of the symbiosis and its maintenance to the impacts on the holobiont development (McFall-Ngai, 2014). The Nematostella anemone, due to its easy maintenance in the lab and sequenced genome, has been developed as a model for metazoan evolution and development and its associated microbiome has been characterized using holobiont metatranscriptomics (Har et al., 2015). S. roscoffensis, whose entire life-cycle can be completed in the lab, serves as a marine model system for photosymbiosis and for the study of physiological processes supported by the tripartite association between the host, microalgae and microbiome such as the production of dimethylsulfoniopropionate (DMSP) (Arboleda et al., 2018). Cassiopea sp. and Exaiptasia sp. are the equivalent of “laboratory rats” for cnidarian research, especially the cnidarian-zooxanthellae symbiosis (Lampert, 2016; Rädecker et al., 2018) and, more recently, as models with the potential of providing a basic functional understanding of cnidarian microbiomes (Herrera et al., 2017).
In parallel to studying model organisms to resolve evolutionary and functional questions, the study of non-model ecologically important species remains essential to provide valuable insights into ecosystem functioning and resilience of key species. There is indisputable evidence that host-microbiome interactions not only impact the holobiont but also underpin the health of some of the most threatened marine ecosystems (Wilkins et al., 2019). Ideally, the knowledge gained from the study of model species and model systems needs to be substantiated in ecologically relevant target species in their native environment (Jaspers et al., 2019). Holobionts are engineers that, through the functional roles provided by their associated microorganisms, have adapted to extreme environments, like mollusks, annelids, and arthropods in hydrothermal vents, invertebrates in intertidal zones or corals in oligotrophic seas (Bang et al., 2018). In coastal environments, some of the key holobionts are corals, sponges, macroalgae, seagrasses, or live in mangroves, and saltmarshes (Trevathan-Tackett et al., 2019). These engineer metaorganisms are at the center of attention when it comes to unraveling the driving forces of ecosystem vulnerability and resilience in the face of increased environmental stressors.
To this date, close to half of the scientific literature utilizing the keyword “holobiont” is centered around corals (47%) (Figure 2). Indeed, the term was first employed in a marine biology context for the description of the animal-dinoflagellate symbiosis in coral reef environments (Rowan, 1998). Shortly after, the concept of “coral holobionts” emerged and quickly spread to describe the coral animal, its algal endosymbionts, and associated microbial community (Little et al., 2004; Wegley et al., 2004). Today, the importance of coral-microbiome interactions is widely recognized and integrated in coral research. In the past decade, the coral holobiont has been the central topic of numerous review and perspective papers (see Supplementary Materials for a list of key holobiont papers) discussing the nature of coral–microbes relationships, the diseases associated with the break-down of those interactions, the implications for the ecosystems, and the stakes surrounding coral holobionts in a context of global change (e.g., Rosenberg et al., 2007; Bourne et al., 2009, 2016; Thompson et al., 2014; Peixoto et al., 2017; Torda et al., 2017; Mera and Bourne, 2018; Van Oppen and Blackall, 2019).
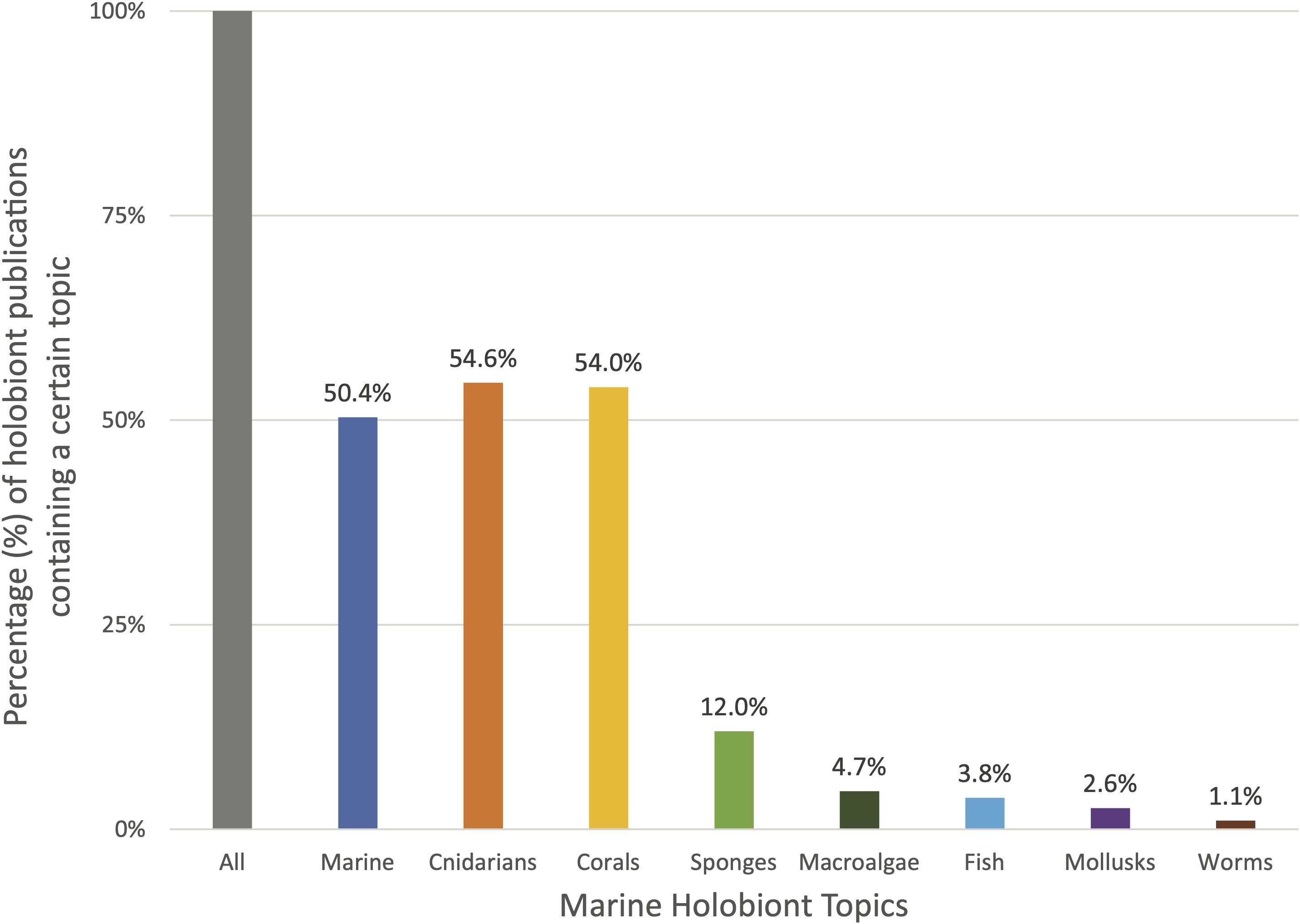
Figure 2. Prevalence of certain marine topics appearing in combination with the term holobiont in the title, abstract, and/or keywords of scientific publications from 1990 to February 2021 (N = 1269). “All” represents the entirety of publications containing the term “holobiont” (100%). The other bars indicate the percentage of publications within this list that contain terms related to a certain marine topic (cnidarians, corals, sponges, etc.). It is important to note that the different topics may overlap in several publications. For example, many publications treat of the topics of coral and sponge holobionts together. Data retrieved from the curated citation and abstract database Scopus on the date 10/03/2021.
Slowly emerging beside the phenomenon of the coral holobiont is the sponge holobiont. Over four decades ago, the early works of Vacelet and Donadey and of Wilkinson first introduced the notion that marine sponges hosted rich microbial communities, and had been doing so over evolutionary significant times (Vacelet and Donadey, 1977; Wilkinson, 1978, 1984). Marine sponges and their incredible microbiome have been recently revisited under the light of the sponge holobiont (Webster and Taylor, 2012; Pita et al., 2018; Kiran et al., 2018; Li et al., 2020). Sponges are vital members of benthic communities across various ecosystems from shallow-water reefs to deep-sea sponge grounds where they act as keystone species by providing a habitat and refuge for other species, thus constituting “biodiversity hotspots” (Bell, 2008; Hogg et al., 2010; Schöttner et al., 2013; Bell et al., 2015). Sponges have been estimated to host up to 40% of their volume worth of associated microorganisms (Webster and Taylor, 2012) and there is increasing evidence that their microbiome underlines key physiological and ecological processes such as nutrient cycling, benthic-pelagic coupling (Bell, 2008), or production of bioactive and antimicrobial compounds which, in addition to their ecological value (Thakur and Singh, 2016), have potential biomedical applications (Sunil Kumar, 2016).
Macroalgae are also key organisms for which microbiome studies are emerging. Indeed, macroalgae are both ecologically and economically important, and their functioning within ecosystems or in an industrial setting is highly dependent on the relationships they maintain with their associated microbiome (Egan et al., 2013; Singh and Reddy, 2016). Several studies have started to investigate the structure and function of seaweed microbial communities and their interplay with environmental change or pathogens (Case et al., 2011; Lachnit et al., 2011; Campbell et al., 2011, 2015; Fernandes et al., 2012; Marzinelli et al., 2015; Singh and Reddy, 2016; Florez et al., 2019).
Ecology and Physiology of Holobiont Organisms
Diversity of Marine Microbes
Microbes in the Global Ocean
The ocean is the largest environment on our planet and hosts the greatest, most dilute microbial system covering the majority of the Earth’s surface and extending to an average depth of 3600 m down to the seafloor (Moran, 2015). Although invisible to the naked eye, marine microbes constitute the majority of the ocean’s biomass with their number reaching into the billions per liter of seawater (Whitman et al., 1998; Moran, 2015). This teeming microscopic world is at the base of marine ecosystem functioning and constitutes a crucial link in trophic food chains through the microbial loop (Pomeroy et al., 2007). Indeed, marine microbes are capable of utilizing molecular-size resources such as drifting proteins, lipids, carbohydrates, and nucleic acids which, in part, are the remains of zooplankton excretion or the cellular innards of lysed plankton cells. In this way, they are reintroducing energy into the food web when higher trophic level species consume them (Pomeroy et al., 2007).
Estimating the diversity, distribution and functional roles of microbes in the global ocean has presented many challenges and brought about debates. Estimates of the total marine microbial diversity range between 106 and 109 bacterial taxa (Pedrós-Alió, 2006) but many questions regarding their specific abundance in various ecosystems, distribution, functional roles, or responses to environmental changes remain unanswered (Salazar and Sunagawa, 2017). The development of sequencing technologies and meta-omics approaches has become central in the quest of linking microbial functions to the microbial community (Moran, 2015). The Tara Ocean expedition running between 2009 and 2013 gathered an international team of scientists who aimed at exploring the morphological, genetic, and functional biodiversity of plankton organisms in the global ocean employing state-of-the-arts technologies such as flow cytometers to monitor viruses, bacteria and small protists, confocal and electron microscopy for 2D/3D imaging of microbial cells as well as HTS for detailed deep phylogenetic rDNA/rRNA tag data and metagenomic/metatranscriptomic functional profiles (Karsenti et al., 2011). This global scale project yielded tremendous amounts of data from dozens of locations around the globe allowing to generate an ocean microbial reference gene catalog with over 40 million non-redundant, mostly novel genomic sequences from viruses, prokaryotes, and picoeukaryotes (Sunagawa et al., 2015). Some of the most abundant taxa across the global ocean belong to members of the Proteobacteria such as the ubiquitous clades SAR11 (Alphaproteobacteria) and SAR86 (Gammaproteobacteria), the Cyanobacteria, the Deferribacteres, and Thaumarchaeota.
Variations in abundance and richness follow a vertical stratification according to changes in light, temperature, and nutrients. Interestingly, the taxonomic and functional richness of mesopelagic and deep-sea microbes is higher than shallow counterparts, whereas cell abundance and potential maximum growth rates decreased with depth (Sunagawa et al., 2015). This pattern is in part explained by the scarcity (mesophotic zone) or total absence of light (aphotic zone) which stimulates the diversification in the utilization of a broad range of alternate energy sources (Ferreira et al., 2014). The deep ocean, and particularly benthic sediments, host microbial engineers who are the driving force of biogeochemical fluxes in the ocean, and, by extension, the Earth’s biogeochemical cycles (Falkowski et al., 2008; Orcutt et al., 2011). Investigating marine microbe diversity and functions across different deep-sea ecosystems is key in order to best predict the impact of climate change and anthropogenic stressors on marine ecosystems. Indeed, for a long time, it was assumed that the deep sea constituted a stable habitat where perturbations were buffered, but recent studies demonstrated that deep-sea microbes are sensitive to changes in temperature, water column oxygenation, pH, availability of trophic resources and food quality with probable scaled-up consequences on marine ecosystems functioning (Corinaldesi, 2015; Danovaro et al., 2016; Sweetman et al., 2017).
Marine microorganisms follow a stratified vertical structure according to physicochemical parameters. The upper ocean presents warmer temperatures and well-mixed surface water, whereas colder temperatures under the thermocline slow the mixing. This results in a limited nutrient supply in the upper thermocline, with nutrients being consumed by phytoplankton for their growth. The upper ocean is depleted in nutrients but is rich in particulate organic carbon accumulating from the phytoplankton. Phytoplankton grow best at a depth, called the “deep chlorophyll maximum” (DCM), where the combination of light and nutrients is optimal (Pierella Karlusich et al., 2020). Eukaryotic phytoplankton such as dinoflagellates and diatoms, and photosynthetic bacteria such as the cyanobacterial genera Prochlorococcus and Synechococcus, constitute the “Earth’s lungs” as they produce significant amounts of oxygen and contribute to almost half of the global net primary production (Field et al., 1998; Partensky et al., 1999). Primary production is highest in coastal and upwelling regions or seasonally at temperate and subpolar latitudes. General trends in phytoplankton community structure show that picocyanobacteria (Prochlorococcus and Synechococcus) dominate in warm, nutrient-poor waters in tropical and subtropical regions, whereas nano- (haptophytes, chlorophytes, pelagophytes, diatoms) and microphytoplankton (diatoms, dinoflagellates) are found year-round in temperate regions and at higher latitudes or upwelling regions respectively (Pierella Karlusich et al., 2020). Much remains to be uncovered regarding the contribution of marine microbes to the functioning of sea-surface ecosystems. However, studies conducting large scale data gathering and meta-analyses are starting to shed light on the spatial patterns and dynamics of microbial communities across large ecosystems such as the Great Barrier Reef (GBR) (Frade et al., 2020). These kinds of studies fall within ecogenomics which is the field of research that aims to understand ecosystem functioning in light of the ecology of microorganisms through the analysis of genomes and metagenomes (Roux et al., 2016; Coutinho et al., 2018).
Archaea and viruses have been discarded from marine research for a long time. However, they have recently been increasingly studied in marine ecosystems, revealing their ubiquity and vital functions in the global ocean. Marine archaea comprise four distinct groups with diverse distributions, physiological and ecological roles. The thaumarchaea are ubiquitous chemolithotrophic organisms that carry a vital role in ammonia oxidation, hence contributing largely to nitrogen and carbon cycling. MGII archaea are heterotrophic and photoheterotrophic organisms whose metabolic diversity remains largely uncharacterized. MGIII archaea, although found throughout the water column, are particularly abundant in the deep-sea where they assumedly perform vital roles in biogeochemical cycling. However, very little is known about the distributions and roles of MGIII and MGIV archaea (Santoro et al., 2019). Viruses are the most abundant biological entities in the ocean and their influence on marine processes and biogeochemical cycles is not to be underestimated (Roux et al., 2016). Indeed, marine viruses are remarkably diverse and can carry a variety of auxiliary metabolic genes encoding vital ecological functions (Breitbart, 2012). Viruses have been both viewed as pathogens potentially causing diseases and as benefactors for the holobiont as they notably exert population control on bacterial and archaeal communities (Sweet and Bythell, 2017). Phages represent the majority of marine viruses. They influence their bacterial hosts in various ways (cell lysis, DNA transfer, manipulation of host metabolism and gene expression, selection for resistance and introduction of novel genetic material), which have radical consequences for marine bacterial assemblages and global biogeochemical cycles (Breitbart, 2012).
Marine Microbiomes
Aside from nutrient cycling and primary productivity in the ocean, marine microbes carry vital functions in host physiology. Microbiomes of several important benthic species have been studied to understand their community structure and function and how they relate to the surrounding environment’s microbial pool (Barott et al., 2011; Ainsworth et al., 2015; Campbell et al., 2015; Jensen et al., 2019; Ziegler et al., 2019; Marchioro et al., 2020). Holobionts present a variable microbiome which differs across distinct individuals of the same species, and a core microbiome which is defined as a group of persistent microbes across all individuals of the same species, for which we can infer a potentially vital role in the holobiont (Hernandez-Agreda et al., 2017; Pita et al., 2018). Studying corals from different locations and different depths allowed to characterize a coral core microbiome which was defined by only several hundred distinct phylotypes in contrast with the much larger global microbial diversity observed in individual corals (Ainsworth et al., 2015). Interestingly, some bacterial phylotypes, identified within the endosymbiotic community, were universal to the core microbiome of coral species that were located in widely separated geographical regions (Ainsworth et al., 2015). Similar results were obtained from sponge holobionts which present species-specific and stable microbiomes across large geographical regions as well as across environmental gradients such as season, depth, and habitat (Pita et al., 2018). Such patterns are also expected in other marine holobionts, such as a kelp species who demonstrated a core microbiome which was influenced by the host health condition rather than its geographical location (Marzinelli et al., 2015). Core microbiomes have also been investigated in scyphozoan jellyfish and some evidence points to the existence of a microbial core-community which is present through different life stages of a same species, highlighting the intrinsic quality of a microbiome for those organisms in their development and evolutionary success (Lee et al., 2018).
Host-associated microbiomes tend to present a clear distinct community composition from the surrounding environment (Barott et al., 2011; Jensen et al., 2019; Marchioro et al., 2020). It was demonstrated that deep-sea corals host highly specific bacterial communities with the most abundant OTUs (operational taxonomic units) only accounting for <0.1% of the surrounding seawater bacterial community (Jensen et al., 2019). Some associated microorganisms called “specialists” are highly specific to certain species with which they maintain a stable partnership, while others, referred to as “generalists,” are abundant across several species and display weaker associations (Jensen et al., 2019). A recent collaborative initiative, the Global Sponge Microbiome Project, aimed at characterizing the microbial diversity in sponges: the most abundant sponge-associated microbes belong to the phyla Proteobacteria (particularly Gamma- and Alphaproteobacteria), Actinobacteria, Chloroflexi, Nitrospirae, Cyanobacteria, Thaumarchaea, and the candidate phylum Poribacteria which is almost exclusively found within sponges (Thomas et al., 2016; Moitinho-Silva et al., 2017; Pita et al., 2018; Orlić, 2019). In corals, the dominant microbial phyla reside within the Proteobacteria (mainly Gamma- and Alphaproteobacteria), Actinobacteria, and Cyanobacteria (Bourne et al., 2016). A recently characterized gammaproteobacterial genus, Endozoicomonas, has attracted attention since the genus (or Endozoicomonas-related bacteria) was reported from various marine invertebrates including corals, sponges, polychetes, ascidians, and mollusks in much higher abundances than in the surrounding sediment or seawater (Shiu and Tang, 2019). In corals, Endozoicomonas dominate the endodermal tissue microbial communities and appear to sustain an intimate and vital relationship with the coral host (Bayer et al., 2013; Neave et al., 2017; Pollock et al., 2018; Marchioro et al., 2020). Indeed, significant decreases of Endozoicomonas abundances in the coral microbiome were observed in diseased corals or following environmental stress (Bourne et al., 2008; Webster et al., 2016). Algae and macroalgae’s microbiomes remain largely understudied but constitute nonetheless a promising field of study. Assessing the microbial diversity of reef algae showed that their microbiome was seemingly more diverse than corals’ microbiomes with a higher abundance of autotrophic bacteria involved in N cycling (Barott et al., 2011). Macroalgae-associated biofilms might provide a niche habitat for microbial indicators of environmental fluctuations such as Firmicutes and Bacteroidota whose ratio increase is indicative of raised nutrient levels and the onset of microbialization in coral reefs (Glasl et al., 2019, 2020). Studying the microbial diversity of certain marine species also reveals crucial links between holobionts, the health of an ecosystem and the threats applied to it. An example is the identification of scyphozoan species-specific microbiomes which, in the case of jellyfish blooms, might influence microbially mediated element cycling in a species-specific manner and act as vectors for pathogens such as Vibrio, Mycoplasma, Ralstonia, Tenacibaculum, Nautella, and Acinetobacter that are harmful to marine organisms and human health (Peng et al., 2021).
Establishment of the Symbiosis, Coevolution, and Adaptive Potential
Transmission, Establishment, and Maintenance
Hosts acquire their symbionts and microbiota either from the surrounding environment through horizontal transfer or from their parents through vertical transfer. Some mixed modes involving both horizontal and vertical transmission or intermediate modes also exist (Bright and Bulgheresi, 2010). Irrespective of the mode of transmission, acquiring a microbiome, establishing and maintaining a microbial community necessitate sophisticated mechanisms involving the host immune system. The transmission of symbionts, whether it is vertically or horizontally, is stressful for both the symbionts and the host. The symbionts have to endure extreme transition in physicochemical conditions (pH, nutrients, trace metals, osmotic and oxidative stress) linked to the characteristics of the niche habitat but also the host developmental hormones and diet. The host’s immune system is greatly challenged as it needs to recognize, accept and incorporate foreign cells. These processes are accompanied by gene expression transitions, chemical and physical alterations to accommodate the newly acquired symbiont (Apprill, 2020).
Some symbionts reside within host cells (endocytosymbionts). Their location is the testimony to a high level of co-dependency and suggests a highly elaborate cellular machinery involved in the maintenance of the relationship (Rosset et al., 2021). Vertical transmission of symbionts requires that the hosts integrate the microbial partners in their reproductive developmental biology through host-mediated mechanisms but also through bacterial molecular mechanisms such as the T3SS (type III secretion system) proteins which allow the bacteria to escape phagocytosis by host immune cells (Dale et al., 2002; Bright and Bulgheresi, 2010). While strict vertical transmission takes place before the progeny leaves the mother, horizontally transmitted symbionts are typically acquired after their dispersal from the parent or after a period of dormancy associated with major developmental morphological modifications (Bright and Bulgheresi, 2010). Horizontal transmission is often mediated through a mucous interface and requires complex recognition mechanisms, most often involving sugar-lectin interactions and cellular surface structures, to select specific symbionts from the environment and avoid pathogen invasions (Bright and Bulgheresi, 2010).
Much remains to be deciphered regarding the specific mechanisms involved in the transmission, establishment and maintenance processes. It is also unclear how prevalent vertical transmission is compared to horizontal transmission for many marine organisms. The coevolutionary theory states that beneficial microbial symbionts are most likely vertically transmitted and that the prevalence of vertical transmission is correlated with the level of dependency of the host on its microbial symbionts (Björk et al., 2019). This has indeed been demonstrated for several symbiotic associations, many of them involving terrestrial insects (Baumann, 2005), some others identified in aquatic organisms such as Hydra (Bathia and Bosch, 2020), in sponges such as Svenzea zeai (Lee et al., 2009) or bivalves containing sulfur-oxidizing chemoautotrophic bacteria (Krueger et al., 1996; Stewart and Cavanaugh, 2006). However, some argue that this does not apply to symbiosis involving a large and diverse microbial community where the vertical transmission of hundreds to thousands of species concomitant with the preservation of their interaction structures and functions is highly improbable (Björk et al., 2019). A recent meta-analysis revealed that, while vertical transmission of microbial symbionts in terrestrial environments seems to prevail, it is remarkably less prevalent in aquatic environments (Russell, 2019). This could arise from the simple fact that water is a conducive medium in which desiccation and osmolarity do not represent a problem, thus encouraging horizontal transmission and host-to-host transfer events (Russell, 2019). Many factors potentially play a role in favoring one mode of transmission over the other. Host dependence and symbiont function are two of them. Indeed, it was demonstrated that host fitness was significantly reduced when nutrient-provisioning, vertically transmitted symbionts were removed (Fisher et al., 2017). This result indicated that functionally important microbes and microbes on which the host is highly dependent tend to be vertically transmitted.
For example, the freshwater Hydra can vertically transfer its photosymbionts, Chlorella, either during asexual reproduction (budding) or through germ-line maternal cells. Although the symbiosis between Hydra and Chlorella indicates a high level of co-dependency, the host is also capable of acquiring the symbionts from the surrounding environment through horizontal acquisition (Bathia and Bosch, 2020). Conversely, the transfer of the luminous Vibrio fisheri symbionts of the bobtail squid Euprymna scolopes occurs exclusively through horizontal acquisition. The processes involved in this acquisition and establishment of the symbiosis have been largely studied, and the squid-Vibrio symbiosis now constitutes a prime model system for horizontal transmission (Nyholm and McFall-Ngai, 2004).
Several studies on sponge microbiomes concluded that vertical transmission was a widespread phenomenon in sponges (Schmitt et al., 2007, 2008; Lee et al., 2009; Webster et al., 2010). 16S rRNA gene tag pyrosequencing allowed to investigate the modes of symbionts transmission in 3 Australian sponge species leading to the identification of sponge-specific symbiont clusters that were either absent or barely detectable in surrounding seawater but were highly enriched in adult sponges and their larvae (Webster et al., 2010). This suggested that vertical transmission and horizontal acquisition of symbionts from the rare seawater microbial community might operate together and both contribute to sponge-microbiome evolution (Webster et al., 2010). Schmitt et al. (2008) argue that “entire microbial consortia are vertically transmitted in sponges” and that additional environmental transfer between adult individuals of the same and even different species might obscure possible signals of co-speciation (Schmitt et al., 2008). A recent study, however, cast doubts on the consistency and faithfulness of vertical transmission in sponges, doubting the feasibility of vertically transmitting multiple microbial species simultaneously and suggesting that complex pathways of indirect transmission also intervene (Björk et al., 2019). These indirect pathways need to be considered during the transmission of diverse and complex microbiomes (Björk et al., 2019).
In corals, the transmission of the algal symbionts can either occur by vertical transmission or horizontal acquisition from the surrounding seawater. Around 85% of scleractinian coral species broadcast spawn gametes into the environment while 15% internally brood their planulae. Spawning corals tend to mainly acquire their Symbiodiniaceae from the environment (∼80%) while brooding corals rely heavily on vertical transmission (∼82%) of the algal symbionts (Baird et al., 2009; Quigley et al., 2017). It would seem that both strategies are employed as their costs and benefits vary depending on environmental conditions. For example, thermal stress conditions could increase the stress in larvae containing high levels of symbionts compared to larvae with low levels of symbionts (Chamberland et al., 2017). To this date, very little is known about the transmission of microbial partners in corals. However, some evidence of vertical transmission exists in brooding corals (Sharp et al., 2012) whose eggs are fertilized inside the polyp and whose larvae are internally brooded. Some corals could also use an intermediate mode of transmission where specific groups of bacteria are released by the parent during spawning and are taken up by their offspring (Ceh et al., 2013). This strategy is not as specific as strict vertical transfer, but more specific than random horizontal acquisition from the surrounding seawater communities (Ceh et al., 2013). The reproductive strategy of the host (brooding vs. spawning) could have an impact on the specificity of certain microbial partners. Spawning corals, which probably acquired microbes from the surrounding seawater, harbored the same Endozoicomonas symbionts across different regions while brooding corals, which release symbiont-rich planula larvae, harbored more specific and geographically distinct Endozoicomonas genotypes (Neave et al., 2017).
Evolution and Adaptation
Arguably the holobiont constitutes a unit of evolutionary selection as it interacts as a whole and manifests adaptation (Roughgarden et al., 2018). A mathematical model showed that, irrespective of symbiont transmission mode, holobiont selection does cause evolutionary changes in holobiont traits (Roughgarden, 2020). This indicates that it believably is an effective evolutionary force, which is something that was contested on the basis that a holobiont’s microbiome is an acquired condition rather than an inherited trait (Roughgarden, 2020). Irrespective of the mode of transmission, hosts and associated microorganisms entertain tight relationships that might intervene in a process of coevolution. Some of the mechanisms that enable symbiotic partners to coevolve include horizontal gene transfer (HGT, the transfer of genetic material between non-mating species, Goldenfeld and Woese, 2007), genome erosion of obligate symbionts (the minimization of genetic information within a genome), strain specificity as well as the growth, reduction and acquisition of novel microbial species from the surrounding environment (Apprill, 2020). In coral genomes, signatures of Symbiodiniaceae and microbial symbiosis are easily identified in the forms of HGT, gene family expansions, and a large collection of innate immunity genes and oxidative stress response genes (Van Oppen and Medina, 2020). These mechanisms of coevolution have the potential to result in longer-term adaptation of the holobiont to environmental stress conditions, leading to a higher resilience (Apprill, 2020). For example, microbial symbionts likely influence immunity evolution of the host through different interactions that will either lead to constraint or relax the selection on immune system maintenance (Gerardo et al., 2020). Commensal microbes that have evolved within the host could contribute to the overall pathogen resistance of the host through competition with pathogens for resources and via direct inhibition. According to some authors, this cooperative immunity, which facilitates the adaptive evolution of non-pathogen-related host traits, is the principal evolutionary advantage provided by the microbiome to their host (McLaren and Callahan, 2020).
However, observing and demonstrating a reciprocal adaptation of diverse microbial lineages with a host lineage is extremely complex. Often, evolutionary studies focus on the inheritance of DNA from parents to offspring. However, this Neo-Darwinian framework, which integrates Mendelian genetics, is challenged by the complexity of symbioses, endosymbioses and host-associated microbiomes (Collens et al., 2019). Increasing evidence shows that the impact of symbionts on host genomes extends beyond the traditional Mendelian views of transmission genetics: symbionts influence host gene expression without changes in DNA sequences. In this way, the hologenome and the co-evolution of host and associated symbionts are an epigenetic phenomenon (Collens et al., 2019). Epigenetic communication between the host and its microbiome, as it was demonstrated in recent studies, is reciprocal and likely involves microRNAs which could regulate gene expression and DNA methylation in the symbiosis partners (Collens et al., 2019). In this context, some authors distinguish between evolutionary adaption, also called adaptation sensu stricto, and adaptation through acclimation, also termed adaptation sensu lato (Webster and Reusch, 2017). The former corresponds to population-level changes in phenotypes due to natural selection and involving genetic change in the form of differing allele frequencies between generations. The later describes transgenerational acclimation in the forms of physiological acclimation, host evolution (often involving epigenetic changes), heritable microbial community changes, and microbial evolution (Webster and Reusch, 2017). Acclimation is a form of phenotypic plasticity describing the adjustment of an organism’s physiology to changing environmental conditions. This is often a reversible phenomenon. In corals, microorganisms evolve much more rapidly than the host and have the potential to rapidly alter the holobiont phenotype via symbionts shuffling and switching, via microbial genetic mutations and HGT. When the microorganisms are vertically transmitted, long-term acclimation of the host is permitted. This is what is called microbiome-mediated transgenerational acclimatization (MMTA) of the coral holobiont (Webster and Reusch, 2017), and this concept could be extended to many marine holobionts. As Amy Apprill points out “symbiosis has played a major role in the evolution of marine organisms and may have driven biological innovation on timescales that are shorter than those of traditional evolutionary responses” (Apprill, 2020). Much remains to be deciphered regarding coevolution and coadaptation of the partners constituting complex holobionts. Mathematical modeling and the study of phylosymbiosis in some key models such as sponges which present a long evolutionary history will help untangle the (co)evolutionary dynamics occurring in complex holobiont systems (O’Brien et al., 2019, 2020).
Functional Roles of Microbiomes
Microbiomes appear to be necessary for their host to thrive as they produce molecules the macroorganism cannot generate by itself. They play a role in nutrient supply, in detoxification, in protection of the holobiont against detrimental microorganisms and in some developmental stages. The well-known examples of sponge and coral holobionts are here explored in details to illustrate the functional roles of microbiomes.
Sponges
Symbionts are necessary for sponge survival as they produce amino acids and vitamins that the animal cannot produce itself (e.g., lysine, histidine, tryptophan, biotin, thiamin, riboflavin, and cobalamin; Fiore et al., 2015). The sponge could collect those needed metabolites by filtration of water, but their associated microbes provide them with these molecules continuously, which ensures them a constant supply.
Genomic studies on phylogenetically divergent species have revealed that their microbiome is capable of nitrogen cycling processes such as denitrification and ammonium oxidation (Fan et al., 2012; Pita et al., 2018). In addition to the direct advantages to the symbionts, the removal of ammonium, nitrate, and nitrite benefits the sponge as those compounds may be harmful if present in large quantities (Hoffmann et al., 2009). It appears that the symbionts can reduce nitrate into nitrite and then into nitric oxide, but do not possess the enzymatic machinery to produce nitrous oxide and finally N2. However, symbionts still use the formers for their growth. Evidence suggests that nitrite could sustain anaerobic growth by its use in acetogenesis. Nitric oxide appears to be used by methanotrophic bacteria to produce nitrogen and oxygen, the latter being used for methane oxidation, thus transforming an anaerobic environment into an aerobic one and maintaining an aerobic respiration even when the sponge is not pumping water (Fan et al., 2012).
Sponge symbionts are able to use molecules produced by their host as a source of carbon and nitrogen, such as creatinine. However, in Xestospongia muta, transcriptomic analyzes showed that the symbionts were using NH3 as a source of nitrogen, while the host was using the catabolism of amino acids. This implies that, if there is any N transfer from the microbiome to the animal, it must be under an organic form (Fiore et al., 2015). In some sponge species, it seems that part of the nitrogen acquired by the animal comes from nitrogen fixation made by cyanobacteria and/or heterotrophic bacteria (Mohamed et al., 2008). This hypothesis is supported by the observation of a low δ15N and the expression of nitrogenase. Different sponge species rely on different ways on their microbiome, especially on photosymbionts, in distinct ways for C and N supply: some use both C and N coming from their symbionts, others use only one of the two forms, and those remaining only feed on particulate organic matter in the filtered water. Those distinctions are perhaps explained by differences in the ability of cyanobacteria to transfer their nutrients to the host, as the microbial species differed between sponges. It is to note that the animal can acclimate to a decline in light, either by feeding heterotrophically or by catching symbiont-derived nutrition, as it still grows under such conditions (Freeman and Thacker, 2011). However, symbionts do not only play a role in the autotrophic supply of carbon. They may play a role in the heterotrophic carbon assimilation, especially in high-microbial abundance sponges (HMA) (Rix et al., 2020). It is hypothesized that, in addition to the surprising direct absorption of dissolved organic matter (DOM) by sponges compared to their well-known particulate organic matter (POM) feeding, microbes also provide them with DOM either by transferring low molecular weight compounds to the animal or by being digested by phagocytosis.
It has been observed in some sponges that both sulfate-reducing bacteria and sulfur-oxidizing bacteria coexist (Gauthier et al., 2016). When oxygen concentration declines, if the animal stops pumping for example, sulfate-reducing bacteria use sulfate as an electron acceptor and produce sulfides, which are toxic above a certain concentration. On the contrary, sulfur-oxidizing microbes prevent sulfides from becoming toxic by oxidizing them to produce carbohydrates from CO2. They can also use reduced sulfur coming from the environment, such as sediments if the sponge is partially covered. Phosphorus is often a limiting nutrient in oligotrophic environments, such as in coral reefs. In addition to the role of free-living microbes, sponge symbionts have been shown to play a significant role in P sequestration. Indeed, they are able to sense the level of inorganic P thanks to a phosphate regulon system, and to create long-chain polyphosphates (Gauthier et al., 2016).
Microbes also produce chemicals that deter predators to feed on their hosts, and secondary metabolites that help the holobiont to compete with other organisms in the ecosystem (Pita et al., 2018). Sponge symbionts are also capable of detoxifying their immediate environment from heavy metals concentrated by the filtering activity of their host (Fan et al., 2012), and to produce antimicrobial compounds to compete with the other microbiome members, which appear to also prevent fouling bacteria from invading the host (Thakur et al., 2004).
In addition to the benefits symbionts offer their host, they have to lose virulence traits and develop protection mechanisms against host defenses, attacks from neighboring microbes and phages (that are concentrated because of the filtering activity of the sponge). As such, genomic analyses often show genes encoding chaperones, membrane proteases and possible evading mechanisms (e.g., ankyrin-repeat proteins) on the one hand and CRISPR or other protecting functions against foreign DNA on the other hand (Gauthier et al., 2016). The production of eukaryotic-like proteins could interfere with eukaryotic protein-protein interactions, especially with proteins from sponge amebocytes to prevent the symbionts from being phagocytized (Burgsdorf et al., 2015). It has already been observed between Legionella pneumophila and their ameba host (Richards et al., 2013). Moreover, symbionts harbor modified antigens (Burgsdorf et al., 2015), or, on the contrary, lack some immune system elicitors such as flagellin (flagella are absent in all sponge microbiome members) (Hayashi et al., 2001; Siegl et al., 2011), enabling them to enter the symbiosis. Hosts appear to recognize their (future) microbial partners via Toll-like receptors, NOD-like receptors and scavenger receptor cysteine-rich (SRCR) family members. Then, some transcription factors (FoxO and NFκβ for example) are upregulated when a microorganism recognized as suitable is present, so that it is maintained in the holobiont. However, the precise mechanisms are not completely understood yet (Pita et al., 2018).
Corals
In corals, symbionts also play a role in nutrient cycling (Bourne et al., 2016). They both produce and degrade carbon molecules. In addition to the well-known role of Symbiodiniaceae in organic carbon supply, cyanobacteria also transfer photosynthates. Microbes catabolize sugars, from simple (e.g., mannose) to complex ones (e.g., cellulose and chitin) (Kimes et al., 2010). Bacteria from Porites astreoides are able to process sugars and proteins that are present in the mucus thereby leading to the hypothesis that they recycle the great amount of energy transferred to this outside layer back to the holobiont (Wegley et al., 2007). Nonetheless, those catabolic properties can have a negative effect, as they trigger the overgrowth of microbes and thus the imbalance of the symbiosis when mono- or polysaccharides are added to the environment (Wegley et al., 2007).
Regarding nitrogen, corals are able to cope with the oligotrophic environment in which they live. Both the animal and Symbiodiniaceae can assimilate ammonium as they both possess glutamine synthetase (GS). Glutamine is transformed into glutamate by glutamine:2-oxoglutarate aminotransferase (GOGAT) in microalgae, and by GOGAT or glutamate dehydrogenase in the animal cells (Pernice et al., 2012). The algae also use nitrate and nitrite reductases to convert NO3 into NH4 (Yellowlees et al., 2008). Diazotrophs from the microbiome can also provide the algal endosymbionts with ammonium obtained from N2 when other nitrogen sources are at least partly depleted (Cardini et al., 2015). However, if NH4 concentration is elevated, it can affect the holobiont fitness by inhibiting the assimilation of carbohydrates by the endosymbionts, and the uptake of nitrate. Thus, mechanisms to prevent an accumulation are needed. Nitrifying and denitrifying microbes are hypothesized to play this role for the holobiont (Siboni et al., 2008). Nitrification could also be used to control Symbiodiniaceae growth as the algae need more energy to use NO3– (Rädecker et al., 2015).
Sulfur is essential for proteins, coenzymes, and metalloproteins and is mostly assimilated under the sulfate form (Sievert et al., 2007). It can be used to synthesize cysteine and methionine, and then DMSP, mainly by algal endosymbionts (Stefels, 2000; Van Alstyne et al., 2006). This product is partly metabolized by bacteria (Raina et al., 2010), and it is hypothesized that some by-products are transferred back to the algae, as it has been observed in other bacterial/algal associations (Amin et al., 2015). Moreover, it is used by a bacterium, Pseudovibrio sp. P12, to produce the antimicrobial tropodithietic acid (TDA) (Raina et al., 2016). Genomic studies have also shown that some bacteria have the potential to reduce sulfate during anaerobic respiration but this process has not been clearly observed yet (Kimes et al., 2010).
Corals also rely on their microbiome to acquire essential vitamins, such as vitamin B12 (Agostini et al., 2012). The prokaryotes do not only provide nutrients and energy to the host; they also protect the holobiont against pathogens either by niche competition or by the production of antimicrobial compounds, such as TDA (Peixoto et al., 2017). However, they are not the only ones to do so. Some viruses, such as the BA3-phage, are known to infect and kill bacterial pathogens, and it is thought that microbiome viruses maintain lysogenic infections of the mutualistic bacteria, which protect them against lytic viruses, whereas opportunistic prokaryotes are susceptible (Sweet and Bythell, 2017).
Focusing on Symbiodiniaceae, evidence supports the idea that they need bacteria to thrive. Firstly, specific bacterial taxa co-localize with the algae inside the coral cells. Secondly, the small genomes of endosymbionts (∼1–5 Gb) imply that they need to associate with other organisms such as the colocalized bacteria (Aranda et al., 2016). Other microalgae-bacteria associations with interesting benefits for the algae (complex compounds degradation, protection, bioavailable iron acquirement) have already been observed. Thirdly, Symbiodiniaceae, when free-living, can form calcified structures with bacteria, called symbiolites which probably protect them against UV and predation and maintain an algal pool available for recruitment by corals (Matthews et al., 2020; Garrido et al., 2021). Similarly, the metamorphosis of some coral larvae appears to be triggered by some microbial biofilms made of bacteria and archaea (Webster et al., 2004).
All the advantages of the microbiome could not be accessible if there was not a robust communication between the host and its microorganisms. Bacteria have been proposed to communicate via quorum-sensing molecules, morphogens, bacterial lipid-based molecules, and vitamins (Van Oppen and Blackall, 2019). Recently, it has been proposed that non-coding RNA could be used as messengers between the members of corals, as those molecules are very old and readable by all types of cells (Leitão et al., 2020). Before any communication or advantage, the host has to build a healthy microbiome. Bacteria are first attracted by chemotaxis toward compounds released by corals and are then categorized as either mutualist or pathogen by the recognition of their microbe-associated molecular patterns (MAMPs). MAMPs are recognized by pattern-recognition receptors (PRRs) but the subsequent mechanisms allowing or not the entry in the symbiosis are not well understood (Bourne et al., 2016).
Marine Holobionts as Drivers of Ecological Processes
Because of all their abilities, microbiomes enable their holobionts to play significant roles for the ecosystems in which they live. The first benefit of microbiomes for ecosystems is their indirect role in key species thriving. With the beneficial roles microbes play for their host (see section “Functional Roles of Microbiomes”), they help engineer species to create the structural and nutritional foundations of their ecosystems (Wilkins et al., 2019), and important processes, such as the sponge loop, to take place. The sponge loop (see Figure 3) consists in the transformation of DOM, unusable by the majority of heterotrophic fauna, to POM with a lower C:N ratio by sponges. Sponges thus provide the biggest source of organic matter available for reef organisms, and part of this process is carried out by their bacteria (Rix et al., 2017). In coral reefs, this production of usable POM appears to be as important as the gross primary production. This is linked to the rapid cell proliferation and turnover of choanocytes, the flagellated cells generating the water flow inside sponges and then absorbing nutritive particles, which releases sponge detritus that form POM. It is hypothesized that microbes having absorbed DOM either translocate low molecular weight compounds or are phagocytosed by choanocytes (Rix et al., 2020). DOM mainly comes from the dissolution of a great part (56 to 80%) of the mucus excreted by corals, and also from algal exudates. Produced POM is then mainly ingested by benthic sponge detritivores, such as ophiuroids, holothuroids, and polychetes. Detritivores are then predated by reef fishes and consequently, sponges enable the transfer of DOM to higher trophic levels (Rix et al., 2018).
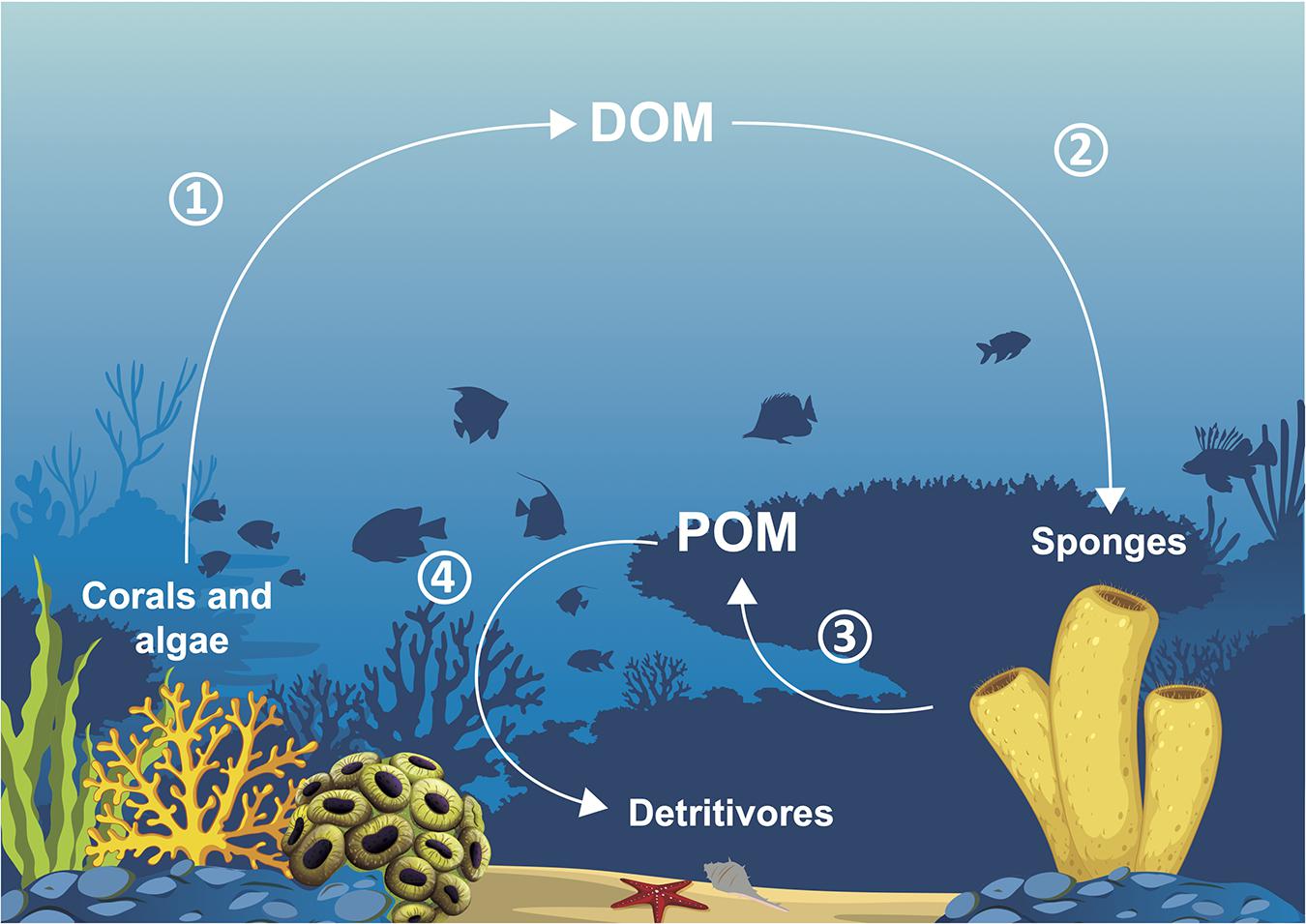
Figure 3. Sponge loop: (1) corals and algae excrete dissolved organic matter (DOM), (2) DOM is taken up by sponges, (3) sponges release detrital particulate organic matter (POM), (4) POM is ingested by detritivores.
Some microbial partners of sponges can deter fishes to feed on their host, such as some cyanobacteria in Lamellodysidea herbacea (Pita et al., 2018). In habitats where predation pressure is high, such capacity has a great impact on the community structure. In addition, the microbiome also helps its host to compete with other organisms, which also shapes the community. For example, bacteria of Terpios hoshinota favor this sponge over corals by the production of cytotoxic secondary metabolites (Pita et al., 2018). It is noteworthy to precise that Epulopiscium gut bacteria of surgeonfish enable this animal to digest polysaccharides from red and brown algae, thus preventing them to overgrow and take the place of corals (Wilkins et al., 2019).
Apart from indirectly providing food, such as the sponge loop, a microbiome can also influence environmental parameters so that they meet the needs of other organisms. For example, aerobic sulfide-oxidizing bacteria associated with benthic bivalves can detoxify anoxic sediments and enable the growth of seagrasses. It is worthy to add that enhancement of microbiome capabilities by biostimulation (addition of limiting nutrients for example) is a promising technique to save ecosystems from being destroyed by hydrocarbons (e.g., oil spill). Coastal microbiomes also appear to be beneficial to humans as they reduce the number of human pathogens in the coastline, probably because of the production of biocides either by the host (seagrass) or by its prokaryotes (Trevathan-Tackett et al., 2019).
Holobionts also play important roles in biogeochemical cycles. It has been demonstrated that sponges release water with more dissolved inorganic nitrogen (DIN) than the water that entered the filtering system. This DIN is either NO3– or NH4+, and the net fluxes are highly variable, depending on many factors such as O2 level and H2S (an inhibitor of both nitrification and denitrification) concentration. The form of released DIN affects the growth of the other organisms, with NH4+ being more easily incorporated than NO3–. In any way, sponges are important producers of DIN in an oligotrophic coral reef environment, as they generate more nitrogen than microbial mats and sediments (Fiore et al., 2013). Moreover, with processes such as denitrification and annamox, sponges also play a significant role in the recycling of nitrogen back to the atmosphere, even more important than the one played by sediments if sponge cover is high (Hoffmann et al., 2009). Both sponges, especially when they host photosynthetic organisms, and corals present important gradients in O2 concentration within their tissues, which favor both aerobic and anaerobic transformations of nitrogen (Moulton et al., 2016). Corals also play a significant role in sulfur cycling. Indeed, their Symbiodiniaceae are great producers of DMSP. 75% of this compound is transformed in methylmercaptopropionate which is then incorporated into bacteria biomass. The remaining 25% is metabolized into dimethyl sulfide (DMS) and acrylic acid. Between 50 and 80% of DMS is used by other types of bacteria, but the remaining 20 to 50% goes back to the atmosphere, where they favor cloud formation and thus, climate cooling (Raina et al., 2009).
Holobionts in Changing Environments
Threats to Marine Holobionts
Climate change, ocean acidification, eutrophication, pollution, and overfishing are the main threats to marine holobionts.
Temperature Rise and Acidification
The rise in temperature has been demonstrated to negatively affect coral holobionts. In addition to the well-known coral bleaching phenomenon (see Weis, 2008; Oakley and Davy, 2018 for more details), the microbiome is also impacted: a rise in temperature decreases the antibacterial capacities of the mucus-associated bacteria (Mouchka et al., 2010). In addition, the healthy microbiome tends to transform into a pathogenic one, especially because of the presence of Vibrio spp. Either those bacteria are replaced by more virulent strains or they undergo metabolic changes that increase the disease potential of the whole holobiont, perhaps because of virally mediated horizontal gene transfer of virulence factors (Thurber et al., 2009). Metagenomic analyses revealed that the virulence genes linked to proteolytic pathways were enhanced with temperature, which means that pathogens tend to lyse holobiont cells during their infection. In addition to bacteria, potentially harmful Microviridae and Fungi related to plant pathogens are also more numerous. Globally, the stress state caused by heat is evidenced by a greater expression of genes related to phenylpropanoids biosynthesis, protection against osmotic and oxidative stress, and repairment of DNA (Littman et al., 2011).
Ocean acidification, which is also caused by climate change, has detrimental effects on corals too. Besides the impairment of coral calcification, which can be decreased by 15–20% (Jokiel et al., 2008), some studies showed that pH could also affect the coral-algal symbiosis. Lower pH induces bleaching, especially under naturally high light, possibly through a disruption of photoprotective mechanisms such as photorespiration or thermal dissipation (Anthony et al., 2008). Acidification also appears to influence coral microbiomes. Morrow and colleagues showed that it was linked to a drop of Endozoicomonas, one of the most abundant bacterial genus in healthy animals, in P. cylindrica and Acropora millepora. Endozoicomonas are thought to provide a great part of the requested nitrogen and sulfur, and to shape the microbiome via signaling molecules and antimicrobial compounds (Morrow et al., 2015).
Regarding sponges, ocean acidification alone could have a beneficial impact on their photoautotrophic symbionts (e.g., Synechococcus sp.) whereas global warming alone would negatively affect those microbes. Indeed, studies demonstrated that cyanobacteria are more abundant on sponges located at natural CO2 seeps than on others subjected to contemporary carbonate concentration, and that an increase in temperature induces a decline of photosynthesis efficiency. When exposed simultaneously to temperature and pH stresses, both photosynthesis and biosynthesis of secondary metabolites involved in chemical defense decrease. Acidification is hypothesized to decrease the ability of the host to regulate its microbiome when exposed to stressors as great inter-individual differences are observed in the composition and the predicted function of the microbes (Lesser et al., 2016). Botté et al. (2019) unveiled a reduction in the transfer of exogenous carbohydrates and amino acids from the animal to the symbionts in CO2 seeps. However, the bicarbonate ion assimilation through the 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle was enhanced in those pH-reduced conditions. In Coelocarteria singaporensis, abundant at CO2 seeps, the host translocates creatine to its symbionts rather than creatinine, thus conserving energy because creatinine degradation uses one ATP to produce two molecules of ammonia whereas creatine degradation also gives two ammonia but does not need any ATP. On the other hand, Stylissa flabelliformis, which is more abundant in control areas, is sensitive to acidification. Its microbiome does not possess the gene battery for creatine metabolism, and thus probably does not receive enough nitrogen from the animal when it is placed in low pH conditions. In addition, these conditions negatively affect the potential of this species to degrade taurine (one of the most abundant free amino acids in sponges) and assimilate sulfur, which could be another explanation of its sensitivity. Hence, all sponge species do not react the same way to ocean acidification (Botté et al., 2019).
Sediments and Nutrients
Another threat to coral reefs is sedimentation and nutrients input. The effects of such stresses on corals are summarized in Bourne et al. (2016). Excess sediment deposition, caused by land practices or dredging for example, has been implicated in diseases expansion, probably because of an immune function reduction, a decrease in energy reserves due to light attenuation, and changes in the microbiome composition. If sediments are especially organic-rich, heterotrophic microorganisms can overgrow, induce the formation of anoxic areas and generate high concentrations of toxic sulfide. More generally, an organic load favors excessive microbial growth, which breaks the balance between the animal and its microbiome, leading to coral death. Corals are also threatened by macroalgae expansion caused by eutrophication and overfishing. The excess of usually limiting nutrients and the reduction of the number of herbivorous fishes induce an overgrowth of algae (Rix et al., 2017). Those organisms harm corals by shading, abrasion and physical injuries, toxic substances release or by attracting corallivorous organisms (Sweet et al., 2013b). In addition, dissolved nutrients excreted by algae can stimulate the overgrowth of coral microbiome and its invasion by opportunistic pathogens (Bourne et al., 2016). Macroalgae were revealed to be reservoirs of pathogenic bacteria and ciliates involved in two coral diseases (white syndrome and yellow band disease) (Sweet et al., 2013b). Sponges seem to tolerate short term nutrients input equivalent to sewage effluent as no visible detrimental effect nor any consequent change in the microbiome were observed in two different species. It is thought that they possibly adapted to increased nutrient concentration as they are abundant on inshore reefs, though precise mechanisms have not been unraveled yet (Simister et al., 2012; Luter et al., 2014). Conversely, sedimentation, even when light intensity was enhanced to counterbalance the opacity caused by sediments, had negative impacts on sponge health and growth. The fitness of those organisms declined even though they did not present significant microbial change and despite their ability to shift from mixotrophy to phototrophy (and vice versa). This decline is perhaps linked to the obligate reduction of food retention when sediments are expelled with mucus in order not to obstruct the aquiferous system (Pineda et al., 2017).
Pathogens and Diseases
Diseases are a major current threat to marine holobionts, especially because they are enhanced by climate change, eutrophication, and overfishing.
Corals
A great difficulty in studying diseases is to identify the causative agent(s), the mode of infection, the genetic basis of virulence, the reservoirs and the vectors, as well as the environmental cofactors (Bourne et al., 2009). Diseases are often correlated to dysbiosis but it is difficult to retrieve the primary agent. Many causative agents have been proposed after microscopic observations or metagenomic analyses of tissue lesions. One has to be careful with those interpretations as causation can only be confirmed by a direct link between the pathogen and the lesion at both gross and cellular levels (Mera and Bourne, 2018). In addition, gross lesions usually used to define a disease can be common to many infections with different pathogens such as for coral white plague disease type II. The symptoms appear in more than 40 species, though a proposed pathogen, Aurantimonas coralicida, has only been demonstrated to fulfill Koch’s postulates with Dichocoenia stokesi (Sunagawa et al., 2009). Those postulates require the isolation of the putative pathogen from every infected host and the apparition of the symptoms on healthy organisms when they are inoculated with this isolate. The same symptoms in Orbicella faveolata were associated with an increase in known coral pathogens and bacteria that had already been isolated from diseased or injured marine invertebrates, but not with A. coralicida. This is an example that adds strength to the hypothesis that disease outbreaks may come from an infection by opportunistic pathogens or the overgrowth of normally commensal bacteria after a weakening of host immune defenses, either caused by a primary infection or an environmental stress (e.g., rise in temperature).
More than 20 coral diseases have already been described, though the causative agents have not been identified for all of them yet. They are summarized in Sheridan et al. (2013). Five of them are illustrated in Figure 4. In some cases, coral bleaching appears to be caused by a pathogen whose virulence has been activated by temperature increase. Two examples are the infections of Oculina patagonica by Vibrio shiloi and Pocillopora damicornis by Vibrio coralliilyticus. The former pathogen expresses a cell-surface adhesin, Toxin P, which blocks the photosynthesis of Symbiodiniaceae, and the superoxide dismutase, the enzyme that initiates the inactivation of superoxide ion, a ROS, in response to heat. The latter pathogen synthetizes an extracellular proteinase which acts as an important virulence factor. The fireworms (Hermodice carunculata) play the role of reservoir for V. shiloi as they feed on corals during summer, extracting the pathogen and keeping it in their body during winter, then re-infecting the cnidarians the next summer (Rosenberg et al., 2007; Sheridan et al., 2013). Bleaching induced by a rise in temperature or by UV also appears to be linked to an activation of latent viruses in some cases (Van Oppen et al., 2009; Sweet and Bythell, 2017). Another Vibrio, Vibrio harveyi, seems to be correlated with the rapid tissue necrosis (Luna et al., 2007).

Figure 4. Examples of coral diseases: (A) gray-patch disease, (B) black band disease, (C) bleaching, (D) white syndrome, (E) brown band disease. Photographs credits: A/Prof. Michael Sweet (A–D) and A/Prof. David Bourne (E).
Black band disease (BBD) is linked to a consortium of pathogens, including cyanobacteria, heterotrophic microbes and sulfate-reducing bacteria (Desulfovibrio spp.). Cyanobacteria provide heterotrophic organisms with organic compounds and oxygen, which stimulate their growth and the production of an anaerobic zone suited for sulfate reduction. The simultaneous action of anoxic environment, cyanobacterial toxins and high sulfide concentration leads to tissue degradation, and finally to the death of the animal (Bourne et al., 2009; Mera and Bourne, 2018; Van Oppen and Blackall, 2019). Gray-patch disease (GPD) presents similarities with BBD: cyanobacteria overgrowth leads to the onset of disease lesions that are subsequently infected by multiple microbes. However, there is no evidence of a rise in sulfate-reducing bacteria. Microbes clustering at the lesions are identical between infected corals, and the competitiveness of this particular assemblage is believed to be the explanation of this great similarity. GPD is less lethal than many other diseases (Sweet et al., 2019). Aspergillosis is an example of disease where the causative agent is difficult to identify. Scientists first thought that this gorgonian illness was linked to the fungus Aspergillus sydowii, before it was defined as an opportunistic pathogen rather than the cause (Mera and Bourne, 2018). Another disease, dark spot syndrome, seems to be linked to a fungus, in addition to bacteria. Indeed, the two most likely pathogens for this disease are a bacteria from the Oscillatoria genus and a fungus closely related to the plant pathogen Rhytisma acerinum (Sweet et al., 2013a). However, one cannot affirm that they are the primary agents given that their correlation with disease lesions does not imply causality. On the contrary, the causative agents of the Caribbean yellow blotch/band disease have been experimentally identified. Four Vibrio spp. extracted from diseased corals triggered the apparition of pale zones when they were inoculated on healthy animals. This loss of color is caused by a lysis of Symbiodiniaceae in the gastroderm. It is not known if those Vibrio species directly induce the degradation of algae or provoke a transformation of normally mutualistic microbes into pathogenic ones. However, whatever the direct action of Vibrio spp., heat increased the disease spread (Cervino et al., 2004). A bacterial causative agent of white pox (WP) was also identified with inoculation experiments (Patterson et al., 2002). The coral Acropora palmata developed irregularly shaped white lesions when it was infected by the human enterobacterium Serratia marcescens and the disease spread more rapidly when temperature was high. White pox was the first disease demonstrated to be caused by human sewage release in the sea.
Another disease inducing the whitening of corals, white band disease (WBD), has also been extensively studied although its causation remains unclear. At first, Vibrio charchariae was proposed to be the primary pathogen (Ritchie and Smith, 1998). However, no subsequent experiment undoubtedly proved it was the case by re-isolating this pathogen from inoculated corals. In 2014, antibiotic treatments allowed to identify 16 microbes (14 bacteria, one archaea and one ciliate) associated with this disease (Sweet et al., 2014). Thirteen of them were identified as secondary pathogens because their elimination with antibiotics did not induce the complete cessation of the disease, whereas three of them (prokaryotes) remain possible primary agents because they could not be eliminated with a specific drug. Gignoux-Wolfsohn et al. (2020) also pinpointed the fact that Rickettsiales-like organisms, which had also been proposed as causative agents, do not increase in abundance on diseased corals. However, they noted that these bacteria, having been observed to multiply in mucocytes and then to rupture them, affect beneficial microorganisms and weaken the holobiont immunity. It is noteworthy to add that Philaster lucinda, the ciliate species, has been associated with WBD and WP (Sutherland et al., 2016). However, in light of the inability of metronidazole, an antiprotozoal agent, to stop WBD lesion progression, this ciliate could be a secondary pathogen, further affecting corals by feeding on tissue and digesting Symbiodiniaceae (Sweet et al., 2014). WBD could be one of the diseases grouped under the name “white syndrome” (WS). Bacterial assemblages of WS vary geographically, and the bacteria and ciliate involved in WBD are identical to one of them (Sweet and Bythell, 2015). Other ciliates also appear to be correlated with other diseases. For example, ciliates, especially Philaster guamensis, are thought to be primarily responsible for the extension of brown band disease lesions. It is even hypothesized that they are the first to infect the coral, before bacteria such as Arcobacter sp. and Aeromonas sp., and after the animal has been injured by a predator, such as the sea star Acanthaster planci. Another Philaster sp. is also associated to brown jelly syndrome, a disease mainly observed in aquaria (Sweet and Bythell, 2012; Sweet et al., 2012). A fourth ciliate, Halofolliculina corallasia is thought to infect corals already damaged by another disease, bleaching or predation and to give rise to Caribbean ciliate infection (Cróquer et al., 2006). In addition to microorganisms, many virus-like particles have been demonstrated to increase in number simultaneously to several diseases (white plague, yellow band syndrome, white syndrome). It is now thought that they play a role, though one cannot say if they are either primary or secondary agents (Sweet and Bythell, 2017).
Sponges
Many disease symptoms have been observed on sponges for decades and worldwide. Many of them are summarized in Webster (2007). They are suspected to be linked to microbes, but only very few causative agents have been determined. The pathogen causing spongin boring necrosis in Rhopaloeides odorabile was identified to be the Alphaproteobacteria strain NW4327 (Webster et al., 2002; Choudhury et al., 2015). Other diseases appear to be linked with non-bacterial microorganisms or viruses, though Koch’s postulates have not been demonstrated yet. Lesions on a single specimen of Verongia cavernicola have been associated with an adenovirus-like particle, diseased tissues of many commercial species with unidentified fungi, and Aplysina red band syndrome with a cyanobacterium. In many studies, a disease-state was correlated with a change of the microbiome from specific to more generalist species. Those observations tend to support the hypothesis that sponge diseases are caused by an imbalance in the microbiome that favors the infection by opportunistic or polymicrobial pathogens (Pita et al., 2018). Studying diseases in Porifera is trickier than in Cnidaria because the former is made of massive three-dimensional tissues that hide disease progression more effectively than the latter that presents thin living tissue on the skeleton. Like corals, sponge diseases will increase with climate change, and probably with eutrophication. Temperature raises virulence of pathogens and reduces host immunity and resilience. In addition, enrichment in inorganic nitrogen and phosphorus likely increases pathogens fitness and virulence (Webster, 2007).
Holobiont Responses, Adaptability, and Vulnerability
Corals’ and sponges’ fates are not easy to define with certitude since some arguments support a decline whereas others are in favor of a resistance to current stressors. As addressed above, environmental changes tend to increase diseases outbreaks by enhancing pathogens virulence and/or reducing host resistance. Moreover, immunity needs energy to be maintained. Stresses that affect dietary resources, such as coral bleaching, can force the holobiont to re-allocate energy where it is indispensable, at the expense of defense. Nevertheless, those organisms are able to protect themselves to some extent (Mullen et al., 2004).
Sponges produce many antibacterial, antiviral, anti-inflammatory, herbicidal and anti-fouling compounds. They fight pathogens that enter their filtering system by releasing cytokines and by phagocytizing them. Animal cells also enter into apoptosis if they get infected, and damaged tissues are isolated from the healthy ones by building tissue barriers. Nitric oxide is also used by sponges against detrimental organisms, and it appears that the production of this gas is enhanced by temperature (Webster, 2007). A recent study revealed that some sponge species are able to enhance their capacity to acquire new microbes in order to sustain their normal growth under acidification. This capacity could perhaps help Porifera to acclimate to environmental changes (Pita et al., 2018).
Corals also possess an innate immunity. Firstly, the acidic mucus repels or traps microbes, and is then evacuated by the movement of apical cilia. Secondly, epidermis forms a physical barrier, followed by a cellular defense consisting of amebocytes and their phagocytosis capacity. In addition, this animal is able to produce antibacterial, antifungal and predator-deterrent compounds (Mullen et al., 2004). The microbiome also produces such compounds, competes with injuring microorganisms for space and nutrients, and contains predators of some detrimental organisms (e.g., Halobacteriovorax spp., predators of Vibrio coralliilyticus) (Van Oppen and Blackall, 2019). However, each aspect of this innate immunity is not equally developed in each species (Mullen et al., 2004).
Though corals do not possess an acquired immunity, it has been observed that they become resistant to some pathogens. For example, V. shiloi cannot induce bleaching of Mediterranean O. patagonica anymore. The bacteria can still adhere to the animal and penetrate into the tissues but they are then lysed in the holobiont cells and they are no longer detectable 4 days after inoculation. This gave rise to the “Coral Probiotic Hypothesis.” This concept states that a dynamic relationship exists between the microbiome and the environment, leading to the selection of microbial partners most suited for the environmental conditions and, more importantly, giving the most advantageous holobiont (see section “Key Concepts in Holobiont Studies”). For the example of V. shiloi and O. patagonica, it is thus believed that a member of the microbiome releases materials that induce the lysis of the pathogen. A change in microbiome implies losses and gains. The microorganisms whose abundance decreases do not multiply rapidly enough to counteract the loss rate induced by mucus removal, or are lysed by bacteriophages. Conversely, microbes growing in abundance were either already present and overgrew or came from the surrounding water or from corallivorous animals (Reshef et al., 2006).
It is supposed that viruses, in addition to their role in protecting microbiome (see section “Functional Roles of Microbiomes”), protect Symbiodiniaceae and cyanobacteria (e.g., Synechococcus sp. phages) from photodamage by encoding D1 and D2 proteins, among others (Van Oppen et al., 2009). However, it is important to note that the protection of the microbiome may be reversed under adverse environmental conditions: lysogenic phages may enter lytic cycle while normally lytic viruses attacking foreign microbes may lose their virulence with stressors (Sweet and Bythell, 2017).
A key for corals survival is within-generation plasticity, accompanied by transgenerational plasticity (TGP). TGP has already been observed in the offsprings of Pocillopora damicornis exposed to high temperature and CO2 concentrations, though it is not known if this adaptation is effective throughout the lifespan of F1 generation and beyond. TGP would be particularly necessary for brooding species (species whose mother colony incubates larvae) as the offspring is not highly dispersed, and will thus live in conditions similar to the ones in which its parent lives, and because those species have relatively short lifespans. Yet, the majority of corals are broadcast spawners, which disperse their larvae to wider areas. So TGP is unlikely the major driver of coral adaptability (Torda et al., 2017).
Holobiont Studies Open New Perspectives
The holobiont concept in marine research has opened a whole new world of discoveries, challenges and perspectives. The exponential rise in marine holobiont topics in recent years (see “A Short History of a New Concept” and “Holobiont Models in Marine Environments”) has shed light on the importance of marine microorganisms for holobiont health and survival. In addition to the gain in fundamental understanding of marine organisms’ biology and ecology, the holobiont approach is key to developing applied solutions for the protection and conservation of ecologically relevant marine species such as corals and sponges. Assuredly, as high-throughput methods to analyze microbiomes are increasingly accessible and fine-tuned (Miller et al., 2011), they are becoming efficient tools to monitor ecosystem health and to inform conservation (Webster et al., 2018).
Marine Probiotics, Customized Medicine, and Assisted Evolution
Human related-stressors such as the introduction of pathogens, of non-native species, contamination with antimicrobials from offshore farms, the use of antibiotics in conventional aquaculture, discharge of contaminants from power plants, oil or mining activities are directly affecting natural marine microbiomes (Wilkins et al., 2019). Microbiomes have the potential to respond rapidly to changes in environmental conditions, including stressors linked to global climate change. Thus, microbiome dynamics could act as biomarkers and inform conservation practices. Species sensitivity distribution and functional sensitivity distribution should be used in probability models to improve the assessment of ecological risk and, in this way, establish microbial conservation guidelines (Webster et al., 2018). Some key areas for future research are the elaboration of microbiome health indices for threatened species and the identification of microbial species and functions that could be used as biomarkers of host’s health and disease (West et al., 2019). In addition, characterizing microbiome taxonomic composition and functional diversity in organisms subjected to non-optimal environmental conditions is paramount to identify the important microbial functional roles involved in the maintenance of the host’s resilience (Wilkins et al., 2019). The strategies using microorganisms to promote marine holobionts’ health remain in their infancy but are, nonetheless, promising. They include direct bioaugmentation by enriching the environment with specific microbes and biostimulation of specific metabolisms involved in host resistance and recovery (Wilkins et al., 2019). Microbiome research in threatened animal species from all marine and terrestrial ecosystems is of the utmost importance as it is key to the development of novel conservation practices (West et al., 2019).
The conservation of coral reefs is amongst the most pressing environmental issues of our generation and is an active research priority (National Academies of Sciences Engineering and Medecine, 2019). A strategy of prevention consisting in removing the factors contributing to reduced health and to mortality in corals (i.e., mitigating the impacts of climate change and eliminating sources of pollutions) is, unfortunately, overwhelmed by the recurrence and increased frequency of climate-related stress events. Therefore, an active intervention, in the form of restoration of coral reefs, is required in order to ensure these ecosystems survive and continue to exist. While we still need to work on enhancing our knowledge of coral reef health by investigating coral holobionts’ responses to environmental stressors, their genetics, their physiological traits and their microbiome assemblages, we also need to work on applied solutions in the form a customized medicine for corals (Peixoto et al., 2019). This describes a multi-level approach to alleviate compromised health symptoms in corals and to aid coral resilience. Some key topics to be explored in coral customized medicine include the development of effective coral probiotics, of methods to deliver beneficial compounds (e.g., antioxidants, nutritional supplementation) and microorganisms to corals, pathogen control (phage therapy, antibiotics, niche occupation, and pathogen exclusion), formulation of BMC (beneficial microorganisms for corals) cocktails facilitating remediation of pollutants, mitigating local environmental impacts, and genetic manipulation of corals (Peixoto et al., 2019).
Many holobiont conservation strategies now fit within the assisted evolution approach. The concept of assisted evolution was first developed by Madeleine J. H. van Oppen in the context of enhancing coral reef resilience. This consists in practices involving human intervention to accelerate the rate of naturally occurring processes in holobiont’s evolution (Van Oppen et al., 2015; Damjanovic et al., 2017). The aim is to enhance certain attributes to help species adapt to changing environmental conditions faster than they would via natural selection. For example, reciprocal transplantation of corals between thermally distinct environments was employed to stimulate the change from a heat-sensitive coral microbiome to a heat-tolerant community (Ziegler et al., 2017). Corals that harbored a tolerant microbiome bleached less when exposed to temperature stress. Recently, inoculation of corals with beneficial bacteria was performed on two coral species in a laboratory setting and proved to be a success as the coral recruits were enriched with these beneficial microbes (Damjanovic et al., 2019). However, host factors still influenced the final taxonomic composition and diversity of the associated microbiome, and it is highly probable that environmental factors also have an impact. Therefore, the success of this manipulation and the long-term stability of the inoculated taxa remain to be evaluated in situ. The use of probiotics is a recent but increasingly popular trend in aquaculture as it presents many advantages such as the elimination of the use of antibiotics, improved water quality, prevention of disease and enhanced fish immune responses (Chauhan and Singh, 2019). Marine probiotics are marine bacterial strains favoring the growth of beneficial microorganisms within a microbiome, as well as restricting the proliferation of decay or disease causing pathogens (Kim et al., 2012). Probiotics are increasingly considered for use in coral conservation practices. For instance, a recent experiment demonstrated that the inoculation of corals with probiotics significantly lessened the effects of thermal stress by increasing coral resistance to bleaching and to pathogen invasion (Rosado et al., 2019). Probiotics could also be used for disease mitigation (Ritchie, 2006; Alagely et al., 2011) and have been shown to be efficient for bioremediation of oil following an oil spill (Fragoso Ados Santos et al., 2015; Blackall et al., 2020). The observation that corals of a same species are genetically divergent across habitats and that both Symbiodiniaceae and prokaryotic populations differ across habitats, leads scientists to contemplate assisted evolution strategies such as assisted gene flow, artificial cross-breeding or probiotic inoculations for the preservation of coral reefs (Van Oppen et al., 2018).
Although assisted evolution and manipulation of holobionts’ beneficial microbes represent promising perspectives for the conservation of endangered ecosystems such as coral reefs, they also raise numerous practical and ethical questions. We still lack a tremendous amount of knowledge regarding ecophysiological responses of marine holobionts to environmental stressors as well as regarding host–microbiome and host–pathobiome interactions. We also lack the practical experience in such practices. As such, one of the greatest concern is the introduction of dangerous pathogens along with the transplantation of “enhanced holobionts” back to their original environment (Sweet et al., 2017). These pathogens could originate from aquariums or the locations used to grow the genetically- or microbially enhanced holobionts. Although some urgency exists surrounding the conservation of endangered ecosystems such as coral reefs, the use of potentially beneficial conservation tools should not risk to inadvertently inflict greater harm in the long term (Sweet et al., 2017). Aside from the lack of knowledge concerning the consequences of such practices, important ethical questions regarding the feasibility and the desirability of creating anthropogenically enhanced systems can be raised (Van Oppen et al., 2015). Indeed, the use of genetically modified or anthropogenically enhanced organisms is extremely controversial. By doing so, we put ourselves in a position of “designers” of nature which forces us to rethink our conceptualization of what is natural (Filbee-Dexter and Smajdor, 2019). Thus, assisted evolution should be used with care, with knowledge and as a last resort when other options are ruled out. It is important to remind ourselves that our efforts need to be primarily oriented toward preventing future damage, protecting healthy ecosystems and understanding future change before considering anthropogenically modifying natural systems (Filbee-Dexter and Smajdor, 2019).
Biotechnological Potential and Economic Value of Marine Holobionts
Many holobionts are increasingly recognized as having great biotechnological potential. Numerous biologically active compounds have been, and still are, discovered and isolated from sponges and their associated microorganisms (Taylor et al., 2007; Sunil Kumar, 2016). As such, sponges have long been considered to have a high potential for drug discovery such as anti-cancer or anti-inflammatory agent. Some sponge species, such as sponges from the Irciniidae family, could be well suited for mariculture in the aim to harvest bioactive compounds (Hardoim and Costa, 2014). New technologies have allowed to identify such potential in other marine holobionts. For example, soft corals (octocorals) are now considered a potential rich source in bioactive secondary metabolites such as terpenes and terpenoids, steroids or prostanoids with promising drug leads as some of these compounds possess anticancer, anti-inflammatory, or antimicrobial properties (Van De Water et al., 2018). Although these octocorals’ derived biomolecules have not yet been adapted to clinical use, some are already used in skin-care products or are considered for use as natural anti-fouling compounds on ships (Van De Water et al., 2018). Similarly, other marine holobionts whose microbiomes are increasingly studied arouse interest for their potential use for biotechnological applications and drug discovery. For example, some argue that jellyfish microbiomes harbor several microbial species with high biotechnological potential that remains to be explored (Tinta et al., 2019). Tunicates, similarly to sponges, are increasingly studied as a source of compounds with pharmacological interest (Bauermeister et al., 2018). Although these biotechnological applications are highly attractive, most are still at the stages of initial screening and pre-clinical studies. Sustainable harvesting of those biomolecules represents a major challenge as they are usually present in small quantities and many of the microbes involved in their production are uncultivable under laboratory conditions (Van De Water et al., 2018).
Holobionts are ubiquitous in marine environments where they often support vital ecological processes and act as ecosystem engineers. Beside their ecological roles, marine holobionts have an important economic value. This arises from the nature of the ecosystems in which they thrive such as coral reefs, kelp forests or deep-sea sponge grounds. Human populations rely on the health of these systems for provisioning (fisheries, materials), supporting (trophic cycles, nutrient cycling), regulating services (carbon sequestration and storage, moderation of extreme events, erosion prevention and pollution remediation), and cultural services (tourism, recreational, aesthetic, and spiritual benefits) (Millennium Ecosystem Assessment, 2005).
Conclusion
Studies on holobionts have become more and more numerous in the past 20 years, especially with the emergence of HTS technologies as they greatly facilitate the study of taxonomic and functional diversity in the microbiome. Researchers first focused on evolutionary questions, before addressing the implications of such associations for the fitness of the partners and for the ecosystems. Bacteria, but also archaea, fungi and viruses, appear to be necessary for their hosts as they play key roles in nutrition, development, protection and competition, thus having an impact on biogeochemical cycles and on community structure. To this day, microbiome dynamics within holobionts have not been completely unraveled. However, the identification of a core microbiome across individuals of a same species across various locations and environmental conditions represents a significant step in our understanding of holobiont’s associated microbial communities. All the functions microorganisms play for their hosts have certainly not been totally discovered. Questions also remain about the transmission of symbionts to the offspring, their establishment, and their maintenance through some mechanisms of the host immunity acceptance and microbial adaptation to radically different conditions. Microbiome also enhance acclimation and adaptation capacities of their host via symbionts shuffling and switching, microbial genetic mutations and horizontal gene transfer. However, climate change, eutrophication, pollution and overfishing appear to negatively impact ecologically important holobionts such as several sponge species and corals. These adverse conditions tend to reduce immunity and increase the virulence of some pathogens, leading to an increase in diseases outbreaks. In order to counteract these detrimental effects and to keep the irreplaceable roles holobionts play for ecosystem maintenance, as well as their unique biotechnological potential, research is currently carried out to elaborate biostimulation, bioaugmentation and assisted evolution strategies. However, such practices require that careful attention is brought to the ethical and technical questions.
From coral reefs, to the deep sea, marine holobionts are ubiquitous and support vital ecosystem services. The rise in holobiont studies is exponential as microbiomes are increasingly recognized as key players in many physiological and ecological processes. With this, a series of new perspectives spanning over evolutionary, ecological and conservation questions appear, pushing the study of marine holobionts in the ranks of top priorities for ocean health.
Author Contributions
CS and MM contributed to manuscript writing and revision. SR and J-CP contributed to manuscript revision and acquired funding. All the authors contributed to the article and approved the submitted version.
Funding
CS and MM are Belgian F.R.S.-FNRS Research Fellows. This work was supported by funding from the F.R.S.-FNRS (CDR J.0014.18 and J.0168.20) and the publication cost was covered by ICYMARE.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to kindly thank A/Prof. D. Bourne and A/Prof. M. Sweet for providing us with pictures of coral diseases to illustrate this review. We would also like to thank the editor and the reviewers of this manuscript for their constructive comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.698853/full#supplementary-material
References
Agostini, S., Suzuki, Y., Higuchi, T., Casareto, B. E., Yoshinaga, K., Nakano, Y., et al. (2012). Biological and chemical characteristics of the coral gastric cavity. Coral Reefs 31, 147–156. doi: 10.1007/s00338-011-0831-6
Ainsworth, T. D., Krause, L., Bridge, T., Torda, G., Raina, J. B., Zakrzewski, M., et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274. doi: 10.1038/ismej.2015.39
Alagely, A., Krediet, C. J., Ritchie, K. B., and Teplitski, M. (2011). Signaling-mediated cross-talk modulates swarming and biofilm formation in a coral pathogen Serratia marcescens. ISME J. 5, 1609–1620. doi: 10.1038/ismej.2011.45
Amin, S. A., Hmelo, L. R., Van Tol, H. M., Durham, B. P., Carlson, L. T., Heal, K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Anthony, K. R. N., Kline, D. I., Diaz-Pulido, G., Dove, S., and Hoegh-Guldberg, O. (2008). Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. U S A. 105, 17442–17446. doi: 10.1073/pnas.0804478105
Apprill, A. (2020). The Role of Symbioses in the Adaptation and Stress Responses of Marine Organisms. Annu. Rev. Mar. Sci. 2020:010641. doi: 10.1146/annurev-marine-010419-010641
Aranda, M., Li, Y., Liew, Y. J., Baumgarten, S., Simakov, O., Wilson, M. C., et al. (2016). Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6, 1–15. doi: 10.1038/srep39734
Arboleda, E., Hartenstein, V., Martinez, P., Reichert, H., Sen, S., Sprecher, S., et al. (2018). An Emerging System to Study Photosymbiosis, Brain Regeneration, Chronobiology, and Behavior: The Marine Acoel Symsagittifera roscoffensis. BioEssays 2018:201800107. doi: 10.1002/bies.201800107
Arrigo, K. R. (2005). Marine microorganisms and global nutrient cycles. Nature 437, 349–355. doi: 10.1038/nature04159
Baedke, J., Fábregas-Tejeda, A., and Nieves Delgado, A. (2020). The holobiont concept before Margulis. J. Exp. Zool. Part B Mol. Dev. Evolut. 334, 149–155. doi: 10.1002/jez.b.22931
Baird, A. H., Guest, J. R., and Willis, B. L. (2009). Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu. Rev. Ecol. Evolut. Systemat. 40, 551–571. doi: 10.1146/annurev.ecolsys.110308.120220
Bang, C., Dagan, T., Deines, P., Dubilier, N., Duschl, W. J., Fraune, S., et al. (2018). Metaorganisms in extreme environments: do microbes play a role in organismal adaptation? Zoology 127, 1–19. doi: 10.1016/j.zool.2018.02.004
Barott, K. L., Rodriguez-Brito, B., Janouškovec, J., Marhaver, K. L., Smith, J. E., Keeling, P., et al. (2011). Microbial diversity associated with four functional groups of benthic reef algae and the reef-building coral Montastraea annularis. Environ. Microbiol. 13, 1192–1204. doi: 10.1111/j.1462-2920.2010.02419.x
Bathia, J., and Bosch, T. C. G. (2020). “Symbiotic interactions in the holobiont Hydra,” in Cellular Dialogues in the Holobiont, eds T. C. G. Bosch and M. G. Hadfield (Florida,FL: CRC Press), 65–77. doi: 10.1201/9780429277375-5
Bauermeister, A., Branco, P. C., Furtado, L. C., Jimenez, P. C., Costa-Lotufo, L. V., da Cruz, et al. (2018). Tunicates: A model organism to investigate the effects of associated-microbiota on the production of pharmaceuticals. Drug Discove. Today: Dis. Models 2018:008. doi: 10.1016/j.ddmod.2019.08.008
Baumann, P. (2005). Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 2005:121041. doi: 10.1146/annurev.micro.59.030804.121041
Bayer, T., Neave, M. J., Alsheikh-Hussain, A., Aranda, M., Yum, L. K., Mincer, T., et al. (2013). The microbiome of the red sea coral stylophora pistillata is dominated by tissue-associated endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762. doi: 10.1128/AEM.00695-13
Bell, J. J. (2008). The functional roles of marine sponges. Estuar. Coastal Shelf Sci. 79, 341–353. doi: 10.1016/j.ecss.2008.05.002
Bell, J. J., Mcgrath, E., Biggerstaff, A., Bates, T., Cárdenas, C. A., and Bennett, H. (2015). Global conservation status of sponges. Conservat. Biol. 29, 42–53. doi: 10.1111/cobi.12447
Björk, J. R., Díez-Vives, C., Astudillo-García, C., Archie, E. A., and Montoya, J. M. (2019). Vertical transmission of sponge microbiota is inconsistent and unfaithful. Nat. Ecol. Evolut. 3, 1172–1183. doi: 10.1038/s41559-019-0935-x
Blackall, L. L., Dungan, A. M., Hartman, L. M., and Van Oppen, M. J. H. (2020). Probiotics for corals. Microbiol. Austral. 2020:MA20025. doi: 10.1071/MA20025
Bosch, T. C. G. (2012). What Hydra has to say about the role and origin of symbiotic interactions. Biol. Bull. 223, 78–84. doi: 10.1086/BBLv223n1p78
Botté, E. S., Nielsen, S., Abdul Wahab, M. A., Webster, J., Robbins, S., Thomas, T., et al. (2019). Changes in the metabolic potential of the sponge microbiome under ocean acidification. Nat. Commun. 10, 1–10. doi: 10.1038/s41467-019-12156-y
Bourne, D. G., Garren, M., Work, T. M., Rosenberg, E., Smith, G. W., and Harvell, C. D. (2009). Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562. doi: 10.1016/j.tim.2009.09.004
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the Coral Microbiome: Underpinning the Health and Resilience of Reef Ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Breitbart, M. (2012). Marine viruses: Truth or dare. Annu. Rev. Mar. Sci. 4, 425–448. doi: 10.1146/annurev-marine-120709-142805
Bright, M., and Bulgheresi, S. (2010). A complex journey: Transmission of microbial symbionts. Nat. Rev. Microbiol. 8, 218–230. doi: 10.1038/nrmicro2262
Brucker, R. M., and Bordenstein, S. R. (2013). The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia. Science 341, 667–669. doi: 10.1126/science.1240659
Burgsdorf, I., Slaby, B. M., Handley, K. M., Haber, M., Blom, J., Marshall, C. W., et al. (2015). Lifestyle evolution in cyanobacterial symbionts of sponges. MBio 6:15. doi: 10.1128/mBio.00391-15
Campbell, A. H., Harder, T., Nielsen, S., Kjelleberg, S., and Steinberg, P. D. (2011). Climate change and disease: Bleaching of a chemically defended seaweed. Glob. Change Biol. 17, 2958–2970. doi: 10.1111/j.1365-2486.2011.02456.x
Campbell, A. H., Marzinelli, E. M., Gelber, J., and Steinberg, P. D. (2015). Spatial variability of microbial assemblages associated with a dominant habitat-forming seaweed. Front. Microbiol. 6:230. doi: 10.3389/fmicb.2015.00230
Cardini, U., Bednarz, V. N., Naumann, M. S., van Hoytema, N., Rix, L., Foster, R. A., et al. (2015). Functional significance of dinitrogen fixation in sustaining coral productivity under oligotrophic conditions. Proc. R. Soc. B Biol. Sci. 282:20152257. doi: 10.1098/rspb.2015.2257
Case, R. J., Longford, S. R., Campbell, A. H., Low, A., Tujula, N., Steinberg, P. D., et al. (2011). Temperature induced bacterial virulence and bleaching disease in a chemically defended marine macroalga. Environ. Microbiol. 13, 529–537. doi: 10.1111/j.1462-2920.2010.02356.x
Ceh, J., van Keulen, M., and Bourne, D. G. (2013). Intergenerational Transfer of Specific Bacteria in Corals and Possible Implications for Offspring Fitness. Microbial Ecol. 65, 227–231. doi: 10.1007/s00248-012-0105-z
Cervino, J. M., Hayes, R. L., Polson, S. W., Polson, S. C., Goreau, T. J., Martinez, R. J., et al. (2004). Relationship of Vibrio species infection and elevated temperatures to yellow blotch/band disease in caribbean corals. Appl. Environ. Microbiol. 70, 6855–6864. doi: 10.1128/AEM.70.11.6855-6864.2004
Chamberland, V. F., Latijnhouwers, K. R. W., Huisman, J., Hartmann, A. C., and Vermeij, M. J. A. (2017). Costs and benefits of maternally inherited algal symbionts in coral larvae. Proc. R. Soc. B Biol. Sci. 284:20170852. doi: 10.1098/rspb.2017.0852
Chauhan, A., and Singh, R. (2019). Probiotics in aquaculture: a promising emerging alternative approach. Symbiosis 77, 99–113. doi: 10.1007/s13199-018-0580-1
Choudhury, J. D., Pramanik, A., Webster, N. S., Llewellyn, L. E., Gachhui, R., and Mukherjee, J. (2015). The Pathogen of the Great Barrier Reef Sponge Rhopaloeides odorabile Is a New Strain of Pseudoalteromonas agarivorans Containing Abundant and Diverse Virulence-Related Genes. Mar. Biotechnol. 17, 463–478. doi: 10.1007/s10126-015-9627-y
Collens, A., Kelley, E., and Katz, L. A. (2019). The concept of the hologenome, an epigenetic phenomenon, challenges aspects of the modern evolutionary synthesis. J. Exp. Zool. Part B Mol. Dev. Evolut. 332, 349–355. doi: 10.1002/jez.b.22915
Corinaldesi, C. (2015). New perspectives in benthic deep-sea microbial ecology. Front. Mar. Sci. 2:17. doi: 10.3389/fmars.2015.00017
Coutinho, F. H., Gregoracci, G. B., Walter, J. M., Thompson, C. C., and Thompson, F. L. (2018). Metagenomics Sheds Light on the Ecology of Marine Microbes and Their Viruses. Trends Microbiol. 26, 955–965. doi: 10.1016/j.tim.2018.05.015
Cróquer, A., Bastidas, C., and Lipscomb, D. (2006). Folliculinid ciliates: A new threat to Caribbean corals? Dis. Aquat. Organis. 69, 75–78. doi: 10.3354/dao069075
Dale, C., Plague, G. R., Wang, B., Ochman, H., and Moran, N. A. (2002). Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc. Natl. Acad. Sci. U S A. 99, 12397–12402. doi: 10.1073/pnas.182213299
Damjanovic, K., Blackall, L. L., Webster, N. S., and van Oppen, M. J. H. (2017). The contribution of microbial biotechnology to mitigating coral reef degradation. Microbial Biotechnol. 10, 1236–1243. doi: 10.1111/1751-7915.12769
Damjanovic, K., Van Oppen, M. J. H., Menéndez, P., and Blackall, L. L. (2019). Experimental inoculation of coral recruits with marine bacteria indicates scope for microbiome manipulation in Acropora tenuis and Platygyra daedalea. Front. Microbiol. 10:1702. doi: 10.3389/fmicb.2019.01702
Danovaro, R., Molari, M., Corinaldesi, C., and Dell’Anno, A. (2016). Macroecological drivers of archaea and bacteria in benthic deep-sea ecosystems. Sci. Adv. 2:e1500961. doi: 10.1126/sciadv.1500961
De Bary, A. (1879). “Die Erscheinung Der Symbiose,” in Die Erscheinung der Symbiose, Vol. 121, ed. A. D. Bary (Strassburg: Verlag von Karl J. Trübner), doi: 10.1515/9783111471839-002
DeLong, E. F. (2009). The microbial ocean from genomes to biomes. Nature 459, 200–206. doi: 10.1038/nature08059
Dittami, S., Arboleda, E., Auguet, J.-C., Bigalke, A., Briand, E., Cárdenas, P., et al. (2020). A community perspective on the concept of marine holobionts: current status, challenges, and future directions. PeerJ 2020:27519. doi: 10.5281/zenodo.3696771
Douglas, A. E., and Werren, J. H. (2016). Holes in the hologenome: Why host-microbe symbioses are not holobionts. MBio 7:15. doi: 10.1128/mBio.02099-15
Egan, S., Harder, T., Burke, C., Steinberg, P., Kjelleberg, S., and Thomas, T. (2013). The seaweed holobiont: Understanding seaweed-bacteria interactions. FEMS Microbiol. Rev. 37, 462–476. doi: 10.1111/1574-6976.12011
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive earth’s biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Fan, L., Reynolds, D., Liu, M., Stark, M., Kjelleberg, S., Webster, N. S., et al. (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. U S A. 109, E1878–E1887. doi: 10.1073/pnas.1203287109
Fernandes, N., Steinberg, P., Rusch, D., Kjelleberg, S., and Thomas, T. (2012). Community Structure and Functional Gene Profile of Bacteria on Healthy and Diseased Thalli of the Red Seaweed Delisea pulchra. PLoS One 7:e50854. doi: 10.1371/journal.pone.0050854
Ferreira, A. J. S., Siam, R., Setubal, J. C., Moustafa, A., Sayed, A., Chambergo, F. S., et al. (2014). Core microbial functional activities in ocean environments revealed by global metagenomic profiling analyses. PLoS One 9:97338. doi: 10.1371/journal.pone.0097338
Field, C. B., Behrenfeld, M. J., Randerson, J. T., and Falkowski, P. (1998). Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281, 237–240. doi: 10.1126/science.281.5374.237
Filbee-Dexter, K., and Smajdor, A. (2019). Ethics of assisted evolution in marine conservation. Front. Mar. Sci. 6:20. doi: 10.3389/fmars.2019.00020
Fiore, C. L., Baker, D. M., and Lesser, M. P. (2013). Nitrogen Biogeochemistry in the Caribbean Sponge, Xestospongia muta: A Source or Sink of Dissolved Inorganic Nitrogen? PLoS One 8:e72961. doi: 10.1371/journal.pone.0072961
Fiore, C. L., Labrie, M., Jarett, J. K., and Lesser, M. P. (2015). Transcriptional activity of the giant barrel sponge, Xestospongia muta Holobiont: Molecular evidence for metabolic interchange. Front. Microbiol. 6:364. doi: 10.3389/fmicb.2015.00364
Fisher, R. M., Henry, L. M., Cornwallis, C. K., Kiers, E. T., and West, S. A. (2017). The evolution of host-symbiont dependence. Nat. Commun. 8, 1–8. doi: 10.1038/ncomms15973
Florez, J. Z., Camus, C., Hengst, M. B., Marchant, F., and Buschmann, A. H. (2019). Structure of the epiphytic bacterial communities of Macrocystis pyrifera in localities with contrasting nitrogen concentrations and temperature. Algal Res. 44:101706. doi: 10.1016/j.algal.2019.101706
Frade, P. R., Glasl, B., Matthews, S. A., Mellin, C., Serrão, E. A., Wolfe, K., et al. (2020). Spatial patterns of microbial communities across surface waters of the Great Barrier Reef. Commun. Biol. 3, 1–14. doi: 10.1038/s42003-020-01166-y
Fragoso Ados Santos, H., Duarte, G. A. S., Rachid, C. T. D. C., Chaloub, R. M., Calderon, E. N., et al. (2015). Impact of oil spills on coral reefs can be reduced by bioremediation using probiotic microbiota. Sci. Rep. 5:18268. doi: 10.1038/srep18268
Freeman, C. J., and Thacker, R. W. (2011). Complex interactions between marine sponges and their symbiotic microbial communities. Limnol. Oceanogr. 56, 1577–1586. doi: 10.4319/lo.2011.56.5.1577
Garrido, A. G., Machado, L. F., Zilberberg, C., Leite, D. C., and de, A. (2021). Insights into ‘Symbiodiniaceae phycosphere’ in a coral holobiont. Symbiosis 83, 735–733. doi: 10.1007/s13199-020-00735-3
Gauthier, M. E. A., Watson, J. R., and Degnan, S. M. (2016). Draft genomes shed light on the dual bacterial symbiosis that dominates the microbiome of the coral reef sponge Amphimedon queenslandica. Front. Mar. Sci. 3:196. doi: 10.3389/fmars.2016.00196
Gerardo, N. M., Hoang, K. L., and Stoy, K. S. (2020). Evolution of animal immunity in the light of beneficial symbioses. Philosoph. Transact. R. Soc. B Biol. Sci. 375:20190601. doi: 10.1098/rstb.2019.0601
Gignoux-Wolfsohn, S. A., Precht, W. F., Peters, E. C., Gintert, B. E., and Kaufman, L. S. (2020). Ecology, histopathology, and microbial ecology of a white-band disease outbreak in the threatened staghorn coral Acropora cervicornis. Dis. Aquat. Organis. 137, 217–237. doi: 10.3354/dao03441
Glasl, B., Bourne, D. G., Frade, P. R., Thomas, T., Schaffelke, B., and Webster, N. S. (2019). Microbial indicators of environmental perturbations in coral reef ecosystems. Microbiome 7:94. doi: 10.1186/s40168-019-0705-7
Glasl, B., Robbins, S., Frade, P. R., Marangon, E., Laffy, P. W., Bourne, D. G., et al. (2020). Comparative genome-centric analysis reveals seasonal variation in the function of coral reef microbiomes. ISME J. 14, 1435–1450. doi: 10.1038/s41396-020-0622-6
Goldenfeld, N., and Woese, C. (2007). Biology’s next revolution. Nature 445:369. doi: 10.1038/445369a
Goodwin, S., McPherson, J. D., and McCombie, W. R. (2016). Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351. doi: 10.1038/nrg.2016.49
Har, J. Y., Helbig, T., Lim, J. H., Fernando, S. C., Reitzel, A. M., Penn, K., et al. (2015). Microbial diversity and activity in the Nematostella vectensis holobiont: Insights from 16S rRNA gene sequencing, isolate genomes, and a pilot-scale survey of gene expression. Front. Microbiol. 6:818. doi: 10.3389/fmicb.2015.00818
Hardoim, C. C. P., and Costa, R. (2014). Microbial communities and bioactive compounds in marine sponges of the family irciniidae-A review. Mar. Drugs 12, 5089–5122. doi: 10.3390/md12105089
Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103. doi: 10.1038/35074106
Hernandez-Agreda, A., Gates, R. D., and Ainsworth, T. D. (2017). Defining the Core Microbiome in Corals’ Microbial Soup. Trends Microbiol. 25, 125–140. doi: 10.1016/j.tim.2016.11.003
Herrera, M., Ziegler, M., Voolstra, C. R., and Aranda, M. (2017). Laboratory-cultured strains of the sea anemone Exaiptasia reveal distinct bacterial communities. Front. Mar. Sci. 4:115. doi: 10.3389/fmars.2017.00115
Hoffmann, F., Radax, R., Woebken, D., Holtappels, M., Lavik, G., Rapp, H. T., et al. (2009). Complex nitrogen cycling in the sponge Geodia barretti. Environ. Microbiol. 11, 2228–2243. doi: 10.1111/j.1462-2920.2009.01944.x
Hogg, M. M., Tendal, O. S., Conway, K. W., Pomponi, S. A., van Soest, R. W. M., Gutt, J., et al. (2010). Deep-sea Sponge Grounds: Reservoirs of Biodiversity. Cambridge: World Conservation Monitoring Centre.
Jaspers, C., Fraune, S., Arnold, A. E., Miller, D. J., Bosch, T. C. G., and Voolstra, C. R. (2019). Resolving structure and function of metaorganisms through a holistic framework combining reductionist and integrative approaches. Zoology 133, 81–87. doi: 10.1016/j.zool.2019.02.007
Jensen, S., Hovland, M., Lynch, M. D. J., and Bourne, D. G. (2019). Diversity of deep-water coral-associated bacteria and comparison across depth gradients. FEMS Microbiol. Ecol. 95:fiz091. doi: 10.1093/femsec/fiz091
Jokiel, P. L., Rodgers, K. S., Kuffner, I. B., Andersson, A. J., Cox, E. F., and Mackenzie, F. T. (2008). Ocean acidification and calcifying reef organisms: A mesocosm investigation. Coral Reefs 27, 473–483. doi: 10.1007/s00338-008-0380-9
Karsenti, E., Acinas, S. G., Bork, P., Bowler, C., de Vargas, C., Raes, J., et al. (2011). A holistic approach to marine Eco-systems biology. PLoS Biol. 9:e1001177. doi: 10.1371/journal.pbio.1001177
Kim, S. K., Bhatnagar, I., and Kang, K. H. (2012). “Development of Marine Probiotics. Prospects and Approach,” in Advances in Food and Nutrition Research, Vol. 65, ed. S.-K. Kim (Florida, FL: Academic Press Inc), 353–362. doi: 10.1016/B978-0-12-416003-3.00023-8
Kimes, N. E., Van Nostrand, J. D., Weil, E., Zhou, J., and Morris, P. J. (2010). Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ. Microbiol. 12, 541–556. doi: 10.1111/j.1462-2920.2009.02113.x
Kinlan, B. P., and Gaines, S. D. (2003). Propagule dispersal in marine and terrestrial environments: A community perspective. Ecology 2003:0622. doi: 10.1890/01-0622
Kiran, G. S., Sekar, S., Ramasamy, P., Thinesh, T., Hassan, S., Lipton, A. N., et al. (2018). Marine sponge microbial association: Towards disclosing unique symbiotic interactions. Mar. Environ. Res. 2018:017. doi: 10.1016/j.marenvres.2018.04.017
Knight, R., Vrbanac, A., Taylor, B. C., Aksenov, A., Callewaert, C., Debelius, J., et al. (2018). Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16, 410–422. doi: 10.1038/s41579-018-0029-9
Koskella, B., and Bergelson, J. (2020). The study of host–microbiome (co)evolution across levels of selection. Philosop. Transact. R. Soc. B Biol. Sci. 375:20190604. doi: 10.1098/rstb.2019.0604
Krueger, D. M., Gustafson, R. G., and Cavanaugh, C. M. (1996). Vertical transmission of chemoautotrophic symbionts in the bivalve Solemya velum (Bivalvia: Protobranchia). Biol. Bull. 190, 195–202. doi: 10.2307/1542539
Kundu, P., Blacher, E., Elinav, E., and Pettersson, S. (2017). Our Gut Microbiome: The Evolving Inner Self. Cell 171, 1481–1493. doi: 10.1016/j.cell.2017.11.024
Lachnit, T., Meske, D., Wahl, M., Harder, T., and Schmitz, R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665. doi: 10.1111/j.1462-2920.2010.02371.x
Lampert, K. P. (2016). “Cassiopea and its zooxanthellae,” in The Cnidaria, past, present and Future: The World of Medusa and her Sisters, eds S. Goffredo and Z. Dubinsky (Berlin: Springer International Publishing), 415–423. doi: 10.1007/978-3-319-31305-4_26
Langerhans, R. B. (2008). “Coevolution,” in Encyclopedia of Ecology, Five-Volume Set, eds S. E. Jørgensen and B. D. Fath (Amsterdam: Elsevier Inc), 644–648. doi: 10.1016/B978-008045405-4.00471-7
Lee, M. D., Kling, J. D., Araya, R., and Ceh, J. (2018). Jellyfish life stages shape associated microbial communities, while a core microbiome is maintained across all. Front. Microbiol. 9:1534. doi: 10.3389/fmicb.2018.01534
Lee, O. O., Chiu, P. Y., Wong, Y. H., Pawlik, J. R., and Qian, P. Y. (2009). Evidence for vertical transmission of bacterial symbionts from adult to embryo in the Caribbean Sponge Svenzea zeai. Appl. Environ. Microbiol. 75, 6147–6156. doi: 10.1128/AEM.00023-09
Leitão, A. L., Costa, M. C., Gabriel, A. F., and Enguita, F. J. (2020). Interspecies communication in holobionts by non-coding RNA exchange. Int. J. Mol. Sci. 2020:ijms21072333. doi: 10.3390/ijms21072333
Lesser, M. P., Fiore, C., Slattery, M., and Zaneveld, J. (2016). Climate change stressors destabilize the microbiome of the Caribbean barrel sponge, Xestospongia muta. J. Exp. Mar. Biol. Ecol. 475, 11–18. doi: 10.1016/j.jembe.2015.11.004
Li, Z., Hentschel, U., Webster, N., Olson, J., and Häggblom, M. (2020). Editorial: special issue on sponge microbiome. FEMS Microbiol. Ecol. 96:fiaa075. doi: 10.1093/femsec/fiaa075
Lim, S. J., and Bordenstein, S. R. (2020). An introduction to phylosymbiosis. Proc. R. Soc. B Biol. Sci. 2020:2900. doi: 10.1098/rspb.2019.2900
Little, A. F., Van Oppen, M. J. H., and Willis, B. L. (2004). Flexibility in algal endosymbioses shapes growth in reef corals. Science 304, 1492–1494. doi: 10.1126/science.1095733
Littman, R., Willis, B. L., and Bourne, D. G. (2011). Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 3, 651–660. doi: 10.1111/j.1758-2229.2010.00234.x
Luna, G. M., Biavasco, F., and Danovaro, R. (2007). Bacteria associated with the rapid tissue necrosis of stony corals. Environ. Microbiol. 9, 1851–1857. doi: 10.1111/j.1462-2920.2007.01287.x
Luter, H. M., Gibb, K., and Webster, N. S. (2014). Eutrophication has no short-term effect on the Cymbastela stipitata holobiont. Front. Microbiol. 5:216. doi: 10.3389/fmicb.2014.00216
Marchioro, G. M., Glasl, B., Engelen, A. H., Serrão, E. A., Bourne, D. G., Webster, N. S., et al. (2020). Microbiome dynamics in the tissue and mucus of acroporid corals differ in relation to host and environmental parameters. PeerJ 8:e9644. doi: 10.7717/peerj.9644
Margulis, L., and Fester, R. (eds) (1991). Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. Cambridge, MA: MIT Press.
Marzinelli, E. M., Campbell, A. H., Zozaya Valdes, E., Vergés, A., Nielsen, S., Wernberg, T., et al. (2015). Continental-scale variation in seaweed host-associated bacterial communities is a function of host condition, not geography. Environ. Microbiol. 17, 4078–4088. doi: 10.1111/1462-2920.12972
Matthews, J. L., Raina, J. B., Kahlke, T., Seymour, J. R., van Oppen, M. J. H., and Suggett, D. J. (2020). Symbiodiniaceae-bacteria interactions: rethinking metabolite exchange in reef-building corals as multi-partner metabolic networks. Environ. Microbiol. 2020:14918. doi: 10.1111/1462-2920.14918
McFall-Ngai, M. J. (2014). The importance of microbes in animal development: Lessons from the squid-vibrio symbiosis. Annu. Rev. Microbiol. 2014:103654. doi: 10.1146/annurev-micro-091313-103654
McLaren, M. R., and Callahan, B. J. (2020). Pathogen resistance may be the principal evolutionary advantage provided by the microbiome. Philosop. Transact. R. Soc. B Biol. Sci. 375:20190592. doi: 10.1098/rstb.2019.0592
Mera, H., and Bourne, D. G. (2018). Disentangling causation: complex roles of coral-associated microorganisms in disease. Environ. Microbiol. 2018:13958. doi: 10.1111/1462-2920.13958
Millennium Ecosystem Assessment (2005). Ecosystems and Human Well-being: Synthesis. Washington, D.C: Island Press.
Miller, D. J., Ball, E. E., Forêt, S., and Satoh, N. (2011). Coral genomics and transcriptomics - Ushering in a new era in coral biology. J. Exp. Mar. Biol. Ecol. 2011:031. doi: 10.1016/j.jembe.2011.07.031
Mohamed, N. M., Colman, A. S., Tal, Y., and Hill, R. T. (2008). Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ. Microbiol. 10, 2910–2921. doi: 10.1111/j.1462-2920.2008.01704.x
Moitinho-Silva, L., Nielsen, S., Amir, A., Gonzalez, A., Ackermann, G. L., Cerrano, C., et al. (2017). The sponge microbiome project. GigaScience 6, 1–7. doi: 10.1093/gigascience/gix077
Moran, M. A. (2015). The global ocean microbiome. Science 2015:aac8455. doi: 10.1126/science.aac8455
Moran, N. A., and Sloan, D. B. (2015). The Hologenome Concept: Helpful or Hollow? PLoS Biol. 13:e1002311. doi: 10.1371/journal.pbio.1002311
Morrow, K. M., Bourne, D. G., Humphrey, C., Botté, E. S., Laffy, P., Zaneveld, J., et al. (2015). Natural volcanic CO2 seeps reveal future trajectories for host-microbial associations in corals and sponges. ISME J. 9, 894–908. doi: 10.1038/ismej.2014.188
Mouchka, M. E., Hewson, I., and Harvell, C. D. (2010). “Coral-associated bacterial assemblages: Current knowledge and the potential for climate-driven impacts,” in Integrative and Comparative Biology, Vol. 50, (Oxford: Oxford University Press), 662–674. doi: 10.1093/icb/icq061
Moulton, O. M., Altabet, M. A., Beman, J. M., Deegan, L. A., Lloret, J., Lyons, M. K., et al. (2016). Microbial associations with macrobiota in coastal ecosystems: Patterns and implications for nitrogen cycling. Front. Ecol. Environ. 2016:1262. doi: 10.1002/fee.1262
Mullen, K. M., Peters, E. C., and Harvell, C. D. (2004). “Coral Resistance to Disease,” in Coral Health and Disease, eds E. Rosenberg and Y. Loya (Berlin: Springer), 377–399. doi: 10.1007/978-3-662-06414-6_22
National Academies of Sciences Engineering and Medecine (2019). A Research Review of Interventions to Increase the Persistence and Resilience of Coral Reefs. A Research Review of Interventions to Increase the Persistence and Resilience of Coral Reefs. Washington, D.C: The National Academies Press, doi: 10.17226/25279
Neave, M. J., Rachmawati, R., Xun, L., Michell, C. T., Bourne, D. G., Apprill, A., et al. (2017). Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200. doi: 10.1038/ismej.2016.95
Nyholm, S. V., and McFall-Ngai, M. J. (2004). The winnowing: Establishing the squid - Vibrios symbiosis. Nat. Rev. Microbiol. 2004:nrmicro957. doi: 10.1038/nrmicro957
O’Brien, P. A., Tan, S., Yang, C., Frade, P. R., Andreakis, N., Smith, H. A., et al. (2020). Diverse coral reef invertebrates exhibit patterns of phylosymbiosis. ISME J. 14, 2211–2222. doi: 10.1038/s41396-020-0671-x
O’Brien, P. A., Webster, N. S., Miller, D. J., and Bourne, D. G. (2019). Host-Microbe Coevolution: Applying Evidence from Model Systems to Complex Marine Invertebrate Holobionts. MBio 10:18.
Oakley, C. A., and Davy, S. K. (2018). Cell Biology of Coral Bleaching. Coral Bleaching 2018, 189–211. doi: 10.1007/978-3-319-75393-5_8
Orcutt, B. N., Sylvan, J. B., Knab, N. J., and Edwards, K. J. (2011). Microbial Ecology of the Dark Ocean above, at, and below the Seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. doi: 10.1128/mmbr.00039-10
Orlić, S. (2019). “Microbial Diversity of Sponge/Coral Microbiome,” in Symbiotic Microbiomes of Coral Reefs Sponges and Corals, ed. Z. Li (Netherlands: Springer), 29–41. doi: 10.1007/978-94-024-1612-1_3
Oulhen, N., Schulz, B. J., and Carrier, T. J. (2016). English translation of Heinrich Anton de Bary’s 1878 speech, ‘Die Erscheinung der Symbiose’ (‘De la symbiose’). Netherlands: Springer Netherlands.
Partensky, F., Hess, W. R., and Vaulot, D. (1999). Prochlorococcus, a Marine Photosynthetic Prokaryote of Global Significance. Microbiol. Mol. Biol. Rev. 63, 106–127. doi: 10.1128/mmbr.63.1.106-127.1999
Patterson, K. L., Porter, J. W., Ritchie, K. B., Polson, S. W., Mueller, E., Peters, E. C., et al. (2002). The etiology of white pox, a lethal disease of the Caribbean elkhorn coral, Acropora palmata. Proc. Natl. Acad. Sci. U S A. 99, 8725–8730. doi: 10.1073/pnas.092260099
Pedrós-Alió, C. (2006). Marine microbial diversity: can it be determined? Trends Microbiol. 14, 257–263. doi: 10.1016/j.tim.2006.04.007
Peixoto, R. S., Rosado, P. M., Leite, D. C. A., Rosado, A. S., and Bourne, D. G. (2017). Beneficial microorganisms for corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 8:00341. doi: 10.3389/fmicb.2017.00341
Peixoto, R. S., Sweet, M., and Bourne, D. G. (2019). Customized Medicine for Corals. Front. Mar. Sci. 6:686. doi: 10.3389/fmars.2019.00686
Peng, S., Hao, W., Li, Y., Wang, L., Sun, T., Zhao, J., et al. (2021). Bacterial Communities Associated With Four Blooming Scyphozoan Jellyfish: Potential Species-Specific Consequences for Marine Organisms and Humans Health. Front. Microbiol. 12:1104. doi: 10.3389/fmicb.2021.647089
Pernice, M., Meibom, A., Van Den Heuvel, A., Kopp, C., Domart-Coulon, I., Hoegh-Guldberg, O., et al. (2012). A single-cell view of ammonium assimilation in coral-dinoflagellate symbiosis. ISME J. 6, 1314–1324. doi: 10.1038/ismej.2011.196
Pierella Karlusich, J. J., Ibarbalz, F. M., and Bowler, C. (2020). Phytoplankton in the Tara Ocean. Annu. Rev. Mar. Sci. 2020:010706. doi: 10.1146/annurev-marine-010419-010706
Pineda, M. C., Strehlow, B., Sternel, M., Duckworth, A., Haan, J., Den, et al. (2017). Effects of sediment smothering on the sponge holobiont with implications for dredging management. Sci. Rep. 7, 1–15. doi: 10.1038/s41598-017-05243-x
Pita, L., Rix, L., Slaby, B. M., Franke, A., and Hentschel, U. (2018). The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6:46. doi: 10.1186/s40168-018-0428-1
Planes, S., Allemand, D., Agostini, S., Banaigs, B., Boissin, E., Boss, E., et al. (2019). The Tara Pacific expedition—A pan-ecosystemic approach of the “-omics” complexity of coral reef holobionts across the Pacific Ocean. PLoS Biol. 2019:3000483. doi: 10.1371/journal.pbio.3000483
Pollock, F. J., McMinds, R., Smith, S., Bourne, D. G., Willis, B. L., Medina, M., et al. (2018). Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Communicat. 9, 1–13. doi: 10.1038/s41467-018-07275-x
Pomeroy, L. R., le Williams, P. J. B., Azam, F., and Hobbie, J. E. (2007). The microbial loop. Oceanography 20, 28–33. doi: 10.5670/oceanog.2007.45
Quigley, K. M., Willis, B. L., and Bay, L. K. (2017). Heritability of the Symbiodinium community in vertically-and horizontally-transmitting broadcast spawning corals. Sci. Rep. 7, 1–14. doi: 10.1038/s41598-017-08179-4
Rädecker, N., Pogoreutz, C., Voolstra, C. R., Wiedenmann, J., and Wild, C. (2015). Nitrogen cycling in corals: The key to understanding holobiont functioning? Trends Microbiol. 2015:008. doi: 10.1016/j.tim.2015.03.008
Rädecker, N., Raina, J.-B., Pernice, M., Perna, G., Guagliardo, P., Kilburn, M. R., et al. (2018). Using Aiptasia as a model to study metabolic interactions in Cnidarian-Symbiodinium symbioses. Front. Physiol. 9:00214. doi: 10.3389/fphys.2018.00214
Raina, J. B., Dinsdale, E. A., Willis, B. L., and Bourne, D. G. (2010). Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 18, 101–108. doi: 10.1016/j.tim.2009.12.002
Raina, J. B., Tapiolas, D., Motti, C. A., Foret, S., Seemann, T., Tebben, J., et al. (2016). Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. PeerJ 2016:e2275. doi: 10.7717/peerj.2275
Raina, J. B., Tapiolas, D., Willis, B. L., and Bourne, D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501. doi: 10.1128/AEM.02567-08
Reshef, L., Koren, O., Loya, Y., Zilber-Rosenberg, I., and Rosenberg, E. (2006). The Coral Probiotic Hypothesis. Environ. Microbiol. 8, 2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x
Richards, A. M., Von Dwingelo, J. E., Price, C. T., and Kwaik, Y. A. (2013). Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence 2013:24290. doi: 10.4161/viru.24290
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Ritchie, K. B., and Smith, G. W. (1998). Type II white-band disease. Rev. Biol. Trop. 1998, 199–203.
Rix, L., De Goeij, J. M., Van Oevelen, D., Struck, U., Al-Horani, F. A., Wild, C., et al. (2018). Reef sponges facilitate the transfer of coral-derived organic matter to their associated fauna via the sponge loop. Mar. Ecol. Prog. Ser. 589, 85–96. doi: 10.3354/meps12443
Rix, L., de Goeij, J. M., van Oevelen, D., Struck, U., Al-Horani, F. A., Wild, C., et al. (2017). Differential recycling of coral and algal dissolved organic matter via the sponge loop. Funct. Ecol. 31, 778–789. doi: 10.1111/1365-2435.12758
Rix, L., Ribes, M., Coma, R., Jahn, M. T., de Goeij, J. M., van Oevelen, D., et al. (2020). Heterotrophy in the earliest gut: a single-cell view of heterotrophic carbon and nitrogen assimilation in sponge-microbe symbioses. ISME J. 14, 2554–2567. doi: 10.1038/s41396-020-0706-3
Rosado, P. M., Leite, D. C. A., Duarte, G. A. S., Chaloub, R. M., Jospin, G., Nunes, et al. (2019). Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13, 921–936. doi: 10.1038/s41396-018-0323-6
Rosenberg, E., and Zilber-Rosenberg, I. (2018). The hologenome concept of evolution after 10 years. Microbiome 6:78. doi: 10.1186/s40168-018-0457-9
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 2007:nrmicro1635. doi: 10.1038/nrmicro1635
Rosset, S. L., Oakley, C. A., Ferrier-Pagès, C., Suggett, D. J., Weis, V. M., and Davy, S. K. (2021). The Molecular Language of the Cnidarian–Dinoflagellate Symbiosis. Trends Microbiol. 2021:005. doi: 10.1016/j.tim.2020.08.005
Roughgarden, J. (2020). Holobiont Evolution: Mathematical Model with Vertical vs. Horizontal Microbiome Transmission. Philosop. Theory Pract. Biol. 12:002. doi: 10.3998/ptpbio.16039257.0012.002
Roughgarden, J., Gilbert, S. F., Rosenberg, E., Zilber-Rosenberg, I., and Lloyd, E. A. (2018). Holobionts as Units of Selection and a Model of Their Population Dynamics and Evolution. Biol. Theory 13, 44–65. doi: 10.1007/s13752-017-0287-1
Roux, S., Brum, J. R., Dutilh, B. E., Sunagawa, S., Duhaime, M. B., Loy, A., et al. (2016). Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689–693. doi: 10.1038/nature19366
Rowan, R. (1998). Diversity and ecology of zooxanthellae on coral reefs. J. Phycol. 34, 407–417. doi: 10.1046/j.1529-8817.1998.340407.x
Russell, S. L. (2019). Transmission mode is associated with environment type and taxa across bacteria-eukaryote symbioses: A systematic review and meta-analysis. FEMS Microbiol. Lett. 2019:fnz013. doi: 10.1093/femsle/fnz013
Salazar, G., and Sunagawa, S. (2017). Marine microbial diversity. Curr. Biol. 2017:017. doi: 10.1016/j.cub.2017.01.017
Santoro, A. E., Richter, R. A., and Dupont, C. L. (2019). Planktonic marine archaea. Annu. Rev. Mar. Sci. 2019:063141. doi: 10.1146/annurev-marine-121916-063141
Schmitt, S., Angermeier, H., Schiller, R., Lindquist, N., and Hentschel, U. (2008). Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl. Environ. Microbiol. 74, 7694–7708. doi: 10.1128/AEM.00878-08
Schmitt, S., Weisz, J. B., Lindquist, N., and Hentschel, U. (2007). Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl. Environ. Microbiol. 73, 2067–2078. doi: 10.1128/AEM.01944-06
Schöttner, S., Hoffmann, F., Cárdenas, P., Rapp, H. T., Boetius, A., and Ramette, A. (2013). Relationships between Host Phylogeny, Host Type and Bacterial Community Diversity in Cold-Water Coral Reef Sponges. PLoS One 8:0055505. doi: 10.1371/journal.pone.0055505
Sharp, K. H., Distel, D., and Paul, V. J. (2012). Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 6, 790–801. doi: 10.1038/ismej.2011.144
Sheridan, C., Kramarsky-Winter, E., Sweet, M., Kushmaro, A., and Leal, M. C. (2013). Diseases in coral aquaculture: Causes, implications and preventions. Aquaculture 2013:037. doi: 10.1016/j.aquaculture.2013.02.037
Shiu, J.-H., and Tang, S.-L. (2019). “The Bacteria Endozoicomonas: Community Dynamics, Diversity, Genomes, and Potential Impacts on Corals,” in Symbiotic Microbiomes of Coral Reefs Sponges and Corals, ed. Z. Li (Netherlands: Springer Netherlands), 55–67. doi: 10.1007/978-94-024-1612-1_5
Siboni, N., Ben-Dov, E., Sivan, A., and Kushmaro, A. (2008). Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environ. Microbiol. 10, 2979–2990. doi: 10.1111/j.1462-2920.2008.01718.x
Siegl, A., Kamke, J., Hochmuth, T., Piel, J., Richter, M., Liang, C., et al. (2011). Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 5, 61–70. doi: 10.1038/ismej.2010.95
Sievert, S. M., Kiene, R. P., and Schulz-Vogt, H. N. (2007). The Sulfur Cycle. Oceonography 20, 117–123.
Simister, R., Taylor, M. W., Tsai, P., and Webster, N. (2012). Sponge-Microbe Associations Survive High Nutrients and Temperatures. PLoS One 7:e52220. doi: 10.1371/journal.pone.0052220
Simon, J.-C., Marchesi, J. R., Mougel, C., and Selosse, M.-A. (2019). Host-microbiota interactions: From holobiont theory to analysis. Microbiome 7, 619–614. doi: 10.1186/s40168-019-0619-4
Singh, B. K., Liu, H., and Trivedi, P. (2020). Eco-holobiont: A new concept to identify drivers of host-associated microorganisms. Environ. Microbiol. 22, 564–567. doi: 10.1111/1462-2920.14900
Singh, R. P., and Reddy, C. R. K. (2016). Unraveling the functions of the macroalgal microbiome. Front. Microbiol. 6:1488. doi: 10.3389/fmicb.2015.01488
Stefels, J. (2000). Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 43, 183–197. doi: 10.1016/S1385-1101(00)00030-7
Stewart, F. J., and Cavanaugh, C. M. (2006). Bacterial endosymbioses in Solemya (Mollusca: Bivalvia) - Model systems for studies of symbiont-host adaptation. Int. J. General Mol. Microbiol. 90, 343–360. doi: 10.1007/s10482-006-9086-6
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359
Sunagawa, S., Desantis, T. Z., Piceno, Y. M., Brodie, E. L., Desalvo, M. K., Voolstra, C. R., et al. (2009). Bacterial diversity and white Plague disease-associated community changes in the caribbean coral montastraea faveolata. ISME J. 3, 512–521. doi: 10.1038/ismej.2008.131
Sunil Kumar, P. (2016). Remarks on the chemo biological applications of marine sponges. Mar. Sponges Chemicobiol. Biomed. Applicat. 2016, 97–103. doi: 10.1007/978-81-322-2794-6_7
Sutherland, K. P., Berry, B., Park, A., Kemp, D. W., Kemp, K. M., Lipp, E. K., et al. (2016). Shifting white pox aetiologies affecting Acropora palmata in the Florida keys, 1994-2014. Philosop. Transact. R. Soc. B Biol. Sci. 371:20150205. doi: 10.1098/rstb.2015.0205
Sweet, M. J., Bythell, J. C., and Nugues, M. M. (2013a). Algae as Reservoirs for Coral Pathogens. PLoS One 8:0069717. doi: 10.1371/journal.pone.0069717
Sweet, M., Burn, D., Croquer, A., and Leary, P. (2013b). Characterisation of the Bacterial and Fungal Communities Associated with Different Lesion Sizes of Dark Spot Syndrome Occurring in the Coral Stephanocoenia intersepta. PLoS One 8:e62580. doi: 10.1371/journal.pone.0062580
Sweet, M. J., Croquer, A., and Bythell, J. C. (2014). Experimental antibiotic treatment identifies potential pathogens of white band disease in the endangered Caribbean coral Acropora cervicornis. Proc. R. Soc. B Biol. Sci. 281:20140094. doi: 10.1098/rspb.2014.0094
Sweet, M., and Bythell, J. (2012). Ciliate and bacterial communities associated with White Syndrome and Brown Band Disease in reef−building corals. Environ. Microbiol. 14, 2184–2199.
Sweet, M., and Bythell, J. (2015). White Syndrome in Acropora muricata: Nonspecific bacterial infection and ciliate histophagy. Mol. Ecol. 24, 1150–1159. doi: 10.1111/mec.13097
Sweet, M., and Bythell, J. (2017). The role of viruses in coral health and disease. J. Invertebrate Pathol. 147, 136–144. doi: 10.1016/j.jip.2016.12.005
Sweet, M., Burian, A., Fifer, J., Bulling, M., Elliott, D., and Raymundo, L. (2019). Compositional homogeneity in the pathobiome of a new, slow-spreading coral disease. Microbiome 7, 1–14.
Sweet, M., Jones, R., and Bythell, J. (2012). Coral diseases in aquaria and in nature. J. Mar. Biol. Associat. U. K. 2012:S0025315411001688. doi: 10.1017/S0025315411001688
Sweet, M., Ramsey, A., and Bulling, M. (2017). Designer reefs and coral probiotics: great concepts but are they good practice? Biodiversity 2017:1307786. doi: 10.1080/14888386.2017.1307786
Sweetman, A. K., Thurber, A. R., Smith, C. R., Levin, L. A., Mora, C., Wei, C. L., et al. (2017). Major impacts of climate change on deep-sea benthic ecosystems. Elem. Sci. Anth. 2017:203. doi: 10.1525/elementa.203
Taylor, M. W., Radax, R., Steger, D., and Wagner, M. (2007). Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol. Mol. Biol. Rev. 71, 295–347. doi: 10.1128/mmbr.00040-06
Thakur, N. L., and Singh, A. (2016). Chemical ecology of marine sponges. Berlin: Springer, 37–52. doi: 10.1007/978-81-322-2794-6_3
Thakur, N. L., Anil, A. C., and Müller, W. E. G. (2004). Culturable epibacteria of the marine sponge Ircinia fusca: Temporal variations and their possible role in the epibacterial defense of the host. Aquat. Microb. Ecol. 37, 295–304. doi: 10.3354/ame037295
Theis, K. R., Dheilly, N. M., Klassen, J. L., Brucker, R. M., Baines, J. F., Bosch, T. C. G., et al. (2016). Getting the hologenome concept right: An eco-evolutionary framework for hosts and their microbiomes. MSystems 1:16. doi: 10.1128/mSystems.00028-16
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7:11870.
Thompson, J. R., Rivera, H. E., Closek, C. J., and Medina, M. (2014). Microbes in the coral holobiont: Partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 4:00176. doi: 10.3389/fcimb.2014.00176
Thurber, R. V., Willner-Hall, D., Rodriguez-Mueller, B., Desnues, C., Edwards, R. A., Angly, F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x
Tinta, T., Kogovšek, T., Klun, K., Malej, A., Herndl, G. J., and Turk, V. (2019). Jellyfish-associated microbiome in the marine environment: Exploring its biotechnological potential. Mar. Drugs 2019:md17020094. doi: 10.3390/md17020094
Torda, G., Donelson, J. M., Aranda, M., Barshis, D. J., Bay, L., Berumen, M. L., et al. (2017). Rapid adaptive responses to climate change in corals. Nat. Clim. Change 2017:nclimate3374. doi: 10.1038/nclimate3374
Trevathan-Tackett, S. M., Sherman, C. D. H., Huggett, M. J., Campbell, A. H., Laverock, B., Hurtado-McCormick, V., et al. (2019). A horizon scan of priorities for coastal marine microbiome research. Nat. Ecol. Evolut. 3, 1509–1520. doi: 10.1038/s41559-019-0999-7
Vacelet, J., and Donadey, C. (1977). Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30, 301–314. doi: 10.1016/0022-0981(77)90038-7
Van Alstyne, K. L., Schupp, P., and Slattery, M. (2006). The distribution of dimethylsulfoniopropionate in tropical Pacific coral reef invertebrates. Coral Reefs 25, 321–327. doi: 10.1007/s00338-006-0114-9
Van De Water, J. A. J. M., Allemand, D., and Ferrier-Pagès, C. (2018). Host-microbe interactions in octocoral holobionts - recent advances and perspectives. Microbiome 2018, 431–436. doi: 10.1186/s40168-018-0431-6
Van Oppen, M. J. H., and Blackall, L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 2019, 223–224. doi: 10.1038/s41579-019-0223-4
Van Oppen, M. J. H., and Medina, M. (2020). Coral evolutionary responses to microbial symbioses. Philosop. Transact. R. Soc. B Biol. Sci. 375:20190591. doi: 10.1098/rstb.2019.0591
Van Oppen, M. J. H., Bongaerts, P., Frade, P., Peplow, L. M., Boyd, S. E., Nim, H. T., et al. (2018). Adaptation to reef habitats through selection on the coral animal and its associated microbiome. Mol. Ecol. 27, 2956–2971. doi: 10.1111/mec.14763
Van Oppen, M. J. H., Leong, J. A., and Gates, R. D. (2009). Coral-virus interactions: A double-edged sword? Symbiosis 2009:BF03179964. doi: 10.1007/BF03179964
Van Oppen, M. J. H., Oliver, J. K., Putnam, H. M., and Gates, R. D. (2015). Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. 112, 2307–2313. doi: 10.1073/pnas.1422301112
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. N. Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Vanwonterghem, I., and Webster, N. S. (2020). Coral Reef Microorganisms in a Changing Climate. IScience 2020:100972. doi: 10.1016/j.isci.2020.100972
Webster, N. S. (2007). Sponge disease: A global threat? Environ. Microbiol. 9, 1363–1375. doi: 10.1111/j.1462-2920.2007.01303.x
Webster, N. S., and Reusch, T. B. H. (2017). Microbial contributions to the persistence of coral reefs. ISME J. 2017:66. doi: 10.1038/ismej.2017.66
Webster, N. S., and Taylor, M. W. (2012). Marine sponges and their microbial symbionts: Love and other relationships. Environ. Microbiol. 14, 335–346. doi: 10.1111/j.1462-2920.2011.02460.x
Webster, N. S., Negri, A. P., Botté, E. S., Laffy, P. W., Flores, F., Noonan, S., et al. (2016). Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6, 1–9. doi: 10.1038/srep19324
Webster, N. S., Negri, A. P., Webb, R. I., and Hill, R. T. (2002). A spongin-boring α-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 232, 305–309. doi: 10.3354/meps232305
Webster, N. S., Smith, L. D., Heyward, A. J., Watts, J. E. M., Webb, R. I., Blackall, L. L., et al. (2004). Metamorphosis of a Scleractinian Coral in Response to Microbial Biofilms. Appl. Environ. Microbiol. 70, 1213–1221. doi: 10.1128/AEM.70.2.1213-1221.2004
Webster, N. S., Taylor, M. W., Behnam, F., Lücker, S., Rattei, T., Whalan, S., et al. (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12, 2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x
Webster, N. S., Wagner, M., and Negri, A. P. (2018). Microbial conservation in the Anthropocene. Environ. Microbiol. 2018:14124. doi: 10.1111/1462-2920.14124
Wegley, L., Edwards, R., Rodriguez-Brito, B., Liu, H., and Rohwer, F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x
Wegley, L., Yu, Y., Breitbart, M., Casas, V., Kline, D. I., and Rohwer, F. (2004). Coral-associated Archaea. Mar. Ecol. Prog. Ser. 273, 89–96. doi: 10.3354/meps273089
Weis, V. M. (2008). Cellular mechanisms of Cnidarian bleaching: Stress causes the collapse of symbiosis. J. Exp. Biol. 2008:009597. doi: 10.1242/jeb.009597
West, A. G., Waite, D. W., Deines, P., Bourne, D. G., Digby, A., McKenzie, V. J., et al. (2019). The microbiome in threatened species conservation. Biol. Conservat. 2019:016. doi: 10.1016/j.biocon.2018.11.016
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. U S A. 1998:6578. doi: 10.1073/pnas.95.12.6578
Wilkins, L. G. E., Leray, M., O’Dea, A., Yuen, B., Peixoto, R. S., Pereira, T. J., et al. (2019). Host-associated microbiomes drive structure and function of marine ecosystems. PLoS Biol. 17:e3000533. doi: 10.1371/journal.pbio.3000533
Wilkinson, C. R. (1978). Microbial associations in sponges. I. Ecology, physiology and microbial populations of coral reef sponges. Mar. Biol. 49, 161–167. doi: 10.1007/BF00387115
Wilkinson, C. R. (1984). Immunological evidence for the Precambrian origin of bacterial symbioses in marine sponges. Proc. R. Soc. Lon. Biol. Sci. 220, 509–517. doi: 10.1098/rspb.1984.0017
Yellowlees, D., Rees, T. A. V., and Leggat, W. (2008). Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31, 679–694. doi: 10.1111/j.1365-3040.2008.01802.x
Ziegler, M., Grupstra, C. G. B., Barreto, M. M., Eaton, M., BaOmar, J., Zubier, K., et al. (2019). Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Communicat. 10, 10969–10965. doi: 10.1038/s41467-019-10969-5
Ziegler, M., Seneca, F. O., Yum, L. K., Palumbi, S. R., and Voolstra, C. R. (2017). Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Communicat. 8, 1–8. doi: 10.1038/ncomms14213
Keywords: marine holobionts, symbiosis, microorganisms, host-microbes interactions, environmental stress
Citation: Stévenne C, Micha M, Plumier J-C and Roberty S (2021) Corals and Sponges Under the Light of the Holobiont Concept: How Microbiomes Underpin Our Understanding of Marine Ecosystems. Front. Mar. Sci. 8:698853. doi: 10.3389/fmars.2021.698853
Received: 22 April 2021; Accepted: 08 July 2021;
Published: 16 August 2021.
Edited by:
Viola Liebich, Bremen Society for Natural Sciences, GermanyReviewed by:
Carlotta Nonnis Marzano, University of Bari Aldo Moro, ItalyMaria Pia Miglietta, Texas A&M University at Galveston, United States
Copyright © 2021 Stévenne, Micha, Plumier and Roberty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chloé Stévenne, chloe.stevenne@uliege.be; Maud Micha, m.micha@uliege.be
†These authors share first authorship
 Chloé Stévenne
Chloé Stévenne Maud Micha
Maud Micha Jean-Christophe Plumier
Jean-Christophe Plumier Stéphane Roberty
Stéphane Roberty