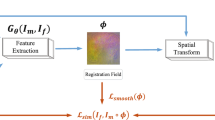
Abstract
Traditional deformable registration methods achieve brilliant results and show strong theoretical support but are computational intensive since they optimize each image pair’s objective function. Recently, supervised learning methods have facilitated fast registration. However, it requires ground truth and does not guarantee a diffeomorphism registration. This paper proposes a new unsupervised learning method Recursive Cascaded Network with a segmentation mask, for two-dimensional medical image registration. Different from the original cascaded network, the network framework into two parts. The first section obtains a pair of image rolls and uses the registration sub-network to predict the deformation vector field from the moving image to the fixed image. The second part introduces anatomical segmentation into the network during training, makes full use of the auxiliary information of the volume, adds an autoencoder to encode the anatomical segmentation, and incorporates it into the learning process of the model in the form of constraints. The local and global ideas are combined to ensure the deformation field’s rationality and improve the distribution. The most important thing is that we propose a formula for calculating the cascaded network’s deformation field used in the test stage to evaluate the relationship between the registration accuracy and the deformation field’s effectiveness. Our experiments show that the system has a better registration effect and less information loss than the current state-of-the-art method. Simultaneously, the cascade method’s accuracy is an improvement at a certain number of layers, and the increase in accuracy needs to sacrifice the effectiveness of the deformation field.










Similar content being viewed by others
References
Fan J, Cao X, Xue Z, Yap P-T, Shen D (2018) Adversarial similarity network for evaluating image alignment in deep learning based registration. In: International conference on medical image computing and computer-assisted intervention. Springer, pp. 739–746
Sokooti H, De Vos B, Berendsen F, Lelieveldt BPF, Išgum I, Staring M (2017) Nonrigid image registration using multi-scale 3d convolutional neural networks. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 232–239
Rohé M-M, Datar M, Heimann T, Sermesant M, Pennec X (2017) Svf-net: learning deformable image registration using shape matching. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 266–274
Lu Z, Yang G, Hua T, Hu L, Kong Y, Tang L, Zhu X, Dillenseger J-L, Shu H, Coatrieux J-L (2019) Unsupervised three-dimensional image registration using a cycle convolutional neural network. In: 2019 IEEE international conference on image processing (ICIP). IEEE, pp 2174–2178
de Vos BD, Berendsen FF, Viergever MA, Staring M, Išgum I (2017) End-to-end unsupervised deformable image registration with a convolutional neural network. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, pp 204–212
Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV (2019) Voxelmorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging 38(8):1788–1800
Zhang J (2018) Inverse-consistent deep networks for unsupervised deformable image registration. arXiv preprint arXiv:1809.03443
Avants BB, Tustison N, Song G (2009) Advanced normalization tools (ants). Insight J 2(365):1–35
Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW (2009) Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29(1):196–205
Hernandez M, Bossa MN, Olmos S (2009) Registration of anatomical images using paths of diffeomorphisms parameterized with stationary vector field flows. Int J Comput Vis 85(3):291–306
Joshi SC, Miller MI (2000) Landmark matching via large deformation diffeomorphisms. IEEE Trans Image Process 9(8):1357–1370
Miller MI, Beg MF, Ceritoglu C, Stark C (2005) Increasing the power of functional maps of the medial temporal lobe by using large deformation diffeomorphic metric mapping. Proc Natl Acad Sci 102(27):9685–9690
Beg MF, Miller MI, Trouvé A, Younes L (2005) Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis 61(2):139–157
Zhang M, Liao R, Dalca AV, Turk EA, Luo J, Grant PE, Golland P (2017) Frequency diffeomorphisms for efficient image registration. In: International conference on information processing in medical imaging. Springer, pp 559–570
Cao Y, Miller MI, Winslow RL, Younes L (2005) Large deformation diffeomorphic metric mapping of vector fields. IEEE Trans Med Imaging 24(9):1216–1230
Ceritoglu C, Oishi K, Li X, Chou M-C, Younes L, Albert M, Lyketsos C, van Zijl PCM, Miller MI, Mori S (2009) Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. Neuroimage 47(2):618–627
Oishi K, Faria A, Jiang H, Li X, Akhter K, Zhang J, Hsu JT, Miller MI, van Zijl PCM, Albert M et al (2009) Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and alzheimer’s disease participants. Neuroimage 46(2):486–499
Ashburner J (2007) A fast diffeomorphic image registration algorithm. Neuroimage 38(1):95–113
Vercauteren T, Pennec X, Perchant A, Ayache N (2009) Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage 45(1):S61–S72
Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV (2018) An unsupervised learning model for deformable medical image registration. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR). IEEE, pp 9252–9260
Shan S, Yan W, Guo X, Chang EI, Fan Y, Xu Y, et al (2017) Unsupervised end-to-end learning for deformable medical image registration. arXiv preprint arXiv:171108608
Li H, Fan Y (2017) Non-rigid image registration using fully convolutional networks with deep self-supervision. arXiv preprint arXiv:170900799
Jaderberg M, Simonyan K, Zisserman A, et al (2015) Spatial transformer networks. In: Advances in neural information processing systems (NIPS), pp 2017–2025
Hu Y, Modat M, Gibson E, Li W, Ghavami N, Bonmati E, Wang G, Bandula S, Moore CM, Emberton M et al (2018) Weakly-supervised convolutional neural networks for multimodal image registration. Med Image Analysis 49:1–13
Hu Y, Modat M, Gibson E, Ghavami N, Bonmati E, Moore CM, Emberton M, Noble JA, Barratt DC, Vercauteren T (2018) Label-driven weakly-supervised learning for multimodal deformable image registration. In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI). IEEE, pp 1070–1074
Zhao S, Dong Y, Chang EI, Xu Y et al (2019) Recursive cascaded networks for unsupervised medical image registration. In: Proceedings of the IEEE international conference on computer vision, pp 10600–10610
Ali S, Rittscher J (2019) Conv2warp: An unsupervised deformable image registration with continuous convolution and warping. In: International workshop on machine learning in medical imaging. Springer, pp 489–497
Zhao S, Lau T, Luo J, Eric I, Chang C, Xu Y (2019) Unsupervised 3d end-to-end medical image registration with volume tweening network. IEEE J Biomed Health Informatics 24(5):1394–1404
Cheng Z, Guo K, Wu C, Shen J, Qu L (2019) U-net cascaded with dilated convolution for medical image registration. In: 2019 Chinese automation congress (CAC). IEEE, pp 3647–3651
Junji S, Shigehiko K, Junpei I, Tsuneo M, Takeshi K, Ken-ichi K, Mitate M, Hiroshi F, Yoshie K, Kunio D (2000) Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am J Roentgenol 174(1):71–74
Candemir S, Jaeger S, Palaniappan K, Musco JP, Singh RK, Xue Z, Karargyris A, Antani S, Thoma G, McDonald CJ (2013) Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans Med Imag 33(2):577–590
Jaeger S, Karargyris A, Candemir S, Folio L, Siegelman J, Callaghan F, Xue Z, Palaniappan K, Singh RK, Antani S et al (2013) Automatic tuberculosis screening using chest radiographs. IEEE Trans Med Imag 33(2):233–245
Rudin LI, Osher S, Fatemi E (1992) Nonlinear total variation based noise removal algorithms. Physica D Nonlinear Phenomena 60(1–4):259–268
Mansilla L, Milone DH, Ferrante E (2020) Learning deformable registration of medical images with anatomical constraints. Neural Netw 124:269–279
Chechik G, Shalit U, Sharma V, Bengio S (2009) An online algorithm for large scale image similarity learning. In: Advances in neural information processing systems (NIPS), pp 306–314
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under Grant 61771322, Grant 61871186, Grant 61971290 and in part by the Fundamental Research Foundation of Shenzhen under Grant JCYJ20190808160815125.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We wish to draw the attention of the Editor to the following facts which may be considered as potential conflicts of interest and to significant financial contributions to this work. We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us. We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing, we confirm that we have followed the regulations of our institutions concerning intellectual property. We understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). He is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. We confirm that we have provided a current, correct email address which is accessible by the Corresponding Author.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
In the process of deriving formula (15) in Sect. 3.3, we found that the following steps can calculate the inverse transformation \(S^{-1}\) of the displacement field:
We can obtain that:
Rights and permissions
About this article
Cite this article
Zou, W., Luo, Y., Cao, W. et al. A cascaded registration network RCINet with segmentation mask. Neural Comput & Applic 33, 16471–16487 (2021). https://doi.org/10.1007/s00521-021-06243-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-06243-9