Abstract
Flowering time (FT) and plant height (PH) are important agronomic traits in soybean. However, their genetic foundations are not fully understood. Thus, in this study, a total of 106,013 single nucleotide polymorphisms in 286 soybean accessions were used to associate with the first and full FT (FT1 and FT2) and PH in 4 environments and their BLUP values using 6 multi-locus genome-wide association study methods. As a result, 38, 43, and 27 stable quantitative trait nucleotides (QTNs) were identified, respectively, for FT1, FT2, and PH across at least 3 methods and/or environments. Among these QTNs for FT1, FT2, and PH, 31, 36, and 21 were found to have significant phenotype differences across 2 alleles; 22, 18, and 13 were consistent with the corresponding loci in previous studies; 13 and 8 genes, with more than average expression level, around 64 FT and 27 PH QTNs were predicted as their corresponding candidate genes. Among these candidate genes, GmPRR3b, and GmGIa for FT, and GmTFL1b for PH were known, while some were new, e.g., GmPHYA4, GmVRN5, GmFPA, and GmSPA1 for FT, and Glyma.02g300200, GmFPA, and Glyma.13g339800 for PH. All the validated QTNs were used to design the best cross-combinations in 2 FT directions. In each FT direction, the best 5 cross-combinations were predicted, such as Heihe 54 × Qincha 1 for early FT, and Yingdejiadou × Wuhuabayuehuang for late FT. This study provides solid foundations for genetic basis, molecular biology, and breeding by design of soybean FT and PH.



Similar content being viewed by others
Data availability
All the data and materials are included in Supplementary Material.
References
Bendix C, Mendoza JM, Stanley DN, Meeley R, Harmon FG (2013) The circadian clock-associated gene gigantea1 affects maize developmental transitions. Plant Cell Environ 36:1379–1390
Bernard RL (1971) Two major genes for time of flowering and maturity in soybeans. Crop Sci 11:242–244
Bonato ER, Vello NA (1999) E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet Mol Biol 22:229–232
Buzzell RI (1971) Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can J Genet Cytol 13:703–707
Buzzell RI, Voldeng HD (1980) Inheritance of insensitivity to long daylength. Soybean Genet News 17:26–29
Chang F, Guo C, Sun F, Zhang J, Wang Z, Kong J, He Q, Sharmin RA, Zhao T (2018) Genome-wide association studies for dynamic plant height and number of nodes on the main stem in summer sowing soybeans. Front Plant Sci 9:1184
Chen L, Nan H, Kong L, Yue L, Yang H, Zhao Q, Fang C, Li H, Cheng Q, Lu S, Kong F, Liu B, Dong L (2020) Soybean AP1 homologs control flowering time and plant height. J Integr Plant Biol 62:1868–1879
Cober ER, Morrison MJ (2010) Regulation of seed yield and agronomic characters by photoperiod sensitivity and growth habit genes in soybean. Theor Appl Genet 120:1005–1012
Cober ER, Voldeng HD (2001) A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci 41:698–701
Cober ER, Molnar SJ, Charette M, Voldeng HD (2010) A new locus for early maturity in soybean. Crop Sci 50:524–527
Cui ZL, Gai JY, Thomas E, Carter TE, Qiu JY, Zhao TJ (1998) The released Chinese soybean cultivars and their pedigree analyses (1923–1995). China Agriculture Press, Beijing
Doyle MR, Bizzell CM, Keller MR, Michaels SD, Song J, Noh YS, Amasino RM (2005) HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J 41:376–385
Fang C, Ma Y, Wu S, Liu Z, Wang Z, Yang R, Hu G, Zhou Z, Yu H, Zhang M, Pan Y, Zhou G, Ren H, Du W, Yan H, Wang Y, Han D, Shen Y, Liu S, Liu T, Zhang J, Qin H, Yuan J, Yuan X, Kong F, Liu B, Li J, Zhang Z, Wang G, Zhu B, Tian Z (2017) Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol 18:161
Fang C, Chen L, Nan H, Kong L, Li Y, Zhang H, Li H, Li T, Tang Y, Hou Z, Dong L, Cheng Q, Lin X, Zhao X, Yuan X, Liu B, Kong F, Lu S (2019) Rapid identification of consistent novel QTLs underlying long-juvenile trait in soybean by multiple genetic populations and genotyping-by-sequencing. Mol Breed 39:80
Fang Y, Liu S, Dong Q, Zhang K, Tian Z, Li X, Li W, Qi Z, Wang Y, Tian X, Song J, Wang J, Yang C, Jiang S, Li WX, Ning H (2020) Linkage analysis and multi-locus genome-wide association studies identify QTNs controlling soybean plant height. Front Plant Sci 11:9
Fernandez MG, Becraft PW, Yin Y, Lübberstedt T (2009) From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci 14:454–461
Fornara F, Montaigu A, Sánchez-Villarreal A, Takahashi Y, Themaat EVL, Huettel B, Davis SJ, Coupland G (2015) The GI–CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J 81:695–706
Funatsuki H, Kawaguchi K, Matsuba S, Sato Y, Ishimoto M (2005) Mapping of QTL associated with chilling tolerance during reproductive growth in soybean. Theor Appl Genet 111:851–861
Gai J, Wang Y, Wu X, Chen S (2007) A comparative study on segregation analysis and QTL mapping of quantitative traits in plants—with a case in soybean. Front Agric China 1:1–7
Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C (2007) The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17:73–78
Higgins RH, Thurber CS, Assaranurak I, Brown PJ (2014) Multiparental Mapping of Plant Height and Flowering Time QTL in Partially Isogenic Sorghum Families. G3 4:1593–1602
Huang J, Li Z, Zhao D (2016) Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci Rep 6:29938
Ikram M, Han X, Zuo JF, Song J, Han CY, Zhang YW, Zhang YM (2020) Identification of QTNs and their candidate genes for 100-seed weight in soybean (Glycine max L.) using multi-locus genome-wide association studies. Genes 11:714
Jing Y, Zhao X, Wang J, Lian M, Teng W, Qiu L, Han Y, Li W (2019) Identification of loci and candidate genes for plant height in soybean (Glycine max) via genome-wide association study. Plant Breed 138:721–732
Kaur S, Atri C, Akhatar J, Mittal M, Kaur R, Banga SS (2021) Genetics of days to flowering, maturity and plant height in natural and derived forms of Brassica rapa L. Theor Appl Genet 134:473–487
Komatsu K, Okuda S, Takahashi M, Matsunaga R, Nakazawa Y (2007) Quantitative trait loci mapping of pubescence density and flowering time of insect-resistant soybean (Glycine max L. Merr.). Genet Mol Biol 30:635–639
Kong F, Nan H, Cao D, Li Y, Wu F, Wang J, Lu S, Yuan X, Cober E, Abe J, Liu B (2014) A new dominant gene E9 conditions early flowering and maturity in soybean. Crop Sci 54:2529–2535
Kuroda Y, Kaga A, Tomooka N, Yano H, Takada Y, Kato S, Vaughan D (2013) QTL affecting fitness of hybrids between wild and cultivated soybeans in experimental fields. Ecol Evol 3:2150–2168
Lee SH, Bailey MA, Mian MR, Carter TE, Ashley DA, Hussey RS, Parrott WA, Boerma HR (1996) Molecular markers associated with soybean plant height, lodging, and maturity across locations. Crop Sci 36:728–735
Li Y, Zhou G, Ma J, Jiang W, Jin L, Zhang Z, Guo Y, Zhang J, Sui Y, Zheng L, Zhang S, Zuo Q, Shi X, Li Y, Zhang W, Hu Y, Kong G, Hong H, Tan B, Song J, Liu Z, Wang Y, Ruan H, Yeung CKL, Liu J, Wang H, Zhang L, Guan R, Wang K, Li W, Chen S, Chang R, Jiang Z, Jackson SA, Li R, Qiu L (2014) De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nature Biotechnol 32:1045–1052
Li Z, Guo Y, Ou L, Hong H, Wang J, Liu Z, Guo B, Zhang L, Qiu L (2018) Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor Appl Genet 131:1001–1016
Li MW, Liu W, Lam HM, Gendron JM (2019) Characterization of two growth period QTLs reveals modification of PRR3 genes during soybean domestication. Plant Cell Physiol 60:407–420
Li C, Li Y, Li Y, Lu H, Hong H, Tian Y, Li H, Zhao T, Zhou X, Liu J, Zhou X, Jackson SA, Liu B, Qiu L (2020a) A domestication-associated gene GmPRR3b regulates the circadian clock and flowering time in soybean. Mol Plant 13:745–759
Li Z, Wang P, You C, Yu J, Zhang X, Yan F, Ye Z, Shen C, Li B, Guo K, Liu N, Thyssen GN, Fang DD, Lindsey K, Zhang X, Wang M, Tu L (2020b) Combined GWAS and eQTL analysis uncovers a genetic regulatory network orchestrating the initiation of secondary cell wall development in cotton. New Phytol 226:1738–1752
Lin X, Liu B, Weller JL, Abe J, Kong F (2020) Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J Integr Plant Biol. https://doi.org/10.1111/jipb.13021
Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180:995–1007
Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, Masuta C, Harada K, Abe J (2010) The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol 153:198–210
Lu S, Dong L, Fang C, Liu S, Kong L, Cheng Q, Chen L, Su T, Nan H, Zhang D, Zhang L, Wang Z, Yang Y, Yu D, Liu X, Yang Q, Lin X, Tang Y, Zhao X, Yang X, Tian C, Xie Q, Li X, Yuan X, Tian Z, Liu B, Weller JL, Kong F (2020) Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nature Genet 52:428–436
Mao T, Li J, Wen Z, Wu T, Wu C, Sun S, Jiang B, Hou W, Li W, Song Q, Wang D, Han T (2017) Association mapping of loci controlling genetic and environmental interaction of soybean flowering time under various photo-thermal conditions. BMC Genomics 18:415
McBlain BA, Bernard RL (1987) A new gene affecting the time of flowering and maturity in soybeans. J Hered 78:160–162
Meuwissen TH, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Mohan M, Nair S, Bhagwat A, Krishna TG, Yano M, Bhatia CR, Sasaki T (1997) Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol Breed 3:87–103
Niu Y, Xu Y, Liu XF, Yang SX, Wei SP, Xie FT, Zhang YM (2013) Association mapping for seed size and shape traits in soybean cultivars. Mol Breed 31:785–794
Oikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M (2004) A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol Biol 5:687–700
Peleman JD, van der Voort JR (2003) Breeding by design. Trends Plant Sci 8:330–334
Ray JD, Hinson K, Mankono JEB, Malo MF (1995) Genetic control of a long-juvenile trait in soybean. Crop Sci 35:1001–1006
Reinprecht Y, Poysa VW, Yu K, Rajcan I, Ablett GR, Pauls KP (2006) Seed and agronomic QTL in low linolenic acid, lipoxygenase-free soybean (Glycine max (L.) Merrill) germplasm. Genome 49:1510–1527
Ren WL, Wen YJ, Dunwell JM, Zhang YM (2018) pKWmEB: integration of Kruskal-Wallis test with empirical Bayes under polygenic background control for multi-locus genome-wide association study. Heredity 120:208–218
Ribaut JM, Hoisington D (1998) Marker-assisted selection: new tools and strategies. Trends Plant Sci 3:236–239
Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M (2002) A mutant gibberellin-synthesis gene in rice. Nature 416:701–702
Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13:1427–1436
Shen Y, Xiang Y, Xu E, Ge X, Li Z (2018) Major co-localized QTL for plant height, branch initiation height, stem diameter, and flowering time in an alien introgression derived Brassica napus DH population. Front Plant Sci 9:390
Tamba CL, Ni YL, Zhang YM (2017) Iterative sure independence screening EM-Bayesian LASSO algorithm for multi-locus genome-wide association studies. PLoS Comput Biol 13:e1005357
Wang R, Liu X, Liang S, Ge Q, Li Y, Shao J, Qi Y, An L, Yu F (2015) A subgroup of MATE transporter genes regulates hypocotyl cell elongation in Arabidopsis. J Exp Bot 66:6327–6343
Wang SB, Feng JY, Ren WL, Huang B, Zhou L, Wen YJ, Zhang J, Dunwell JM, Xu S, Zhang YM (2016) Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci Rep 6:19444
Wang F, Nan H, Chen L, Fang C, Zhang H, Su T, Li S, Cheng Q, Dong L, Liu B, Kong F, Lu S (2019) A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol Breed 39:70
Wang L, Fang C, Liu J, Zhang T, Kou K, Su T, Li S, Chen L, Cheng Q, Dong L, Kong F, Liu B, Lu S (2020) Identification of major QTLs for flowering and maturity in soybean by genotyping-by-sequencing analysis. Mol Breed 40:99
Watanabe S, Xia Z, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K, Harada K (2011) A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188:395–407
Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153:1747–1758
Wen Z, Boyse JF, Song Q, Cregan PB, Wang D (2015) Genomic consequences of selection and genome-wide association mapping in soybean. BMC Genomics 16:671
Wen YJ, Zhang H, Ni YL, Huang B, Zhang J, Feng JY, Wang SB, Dunwell JM, Zhang YM, Wu R (2018) Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief Bioinform 19:700–712
Xia Z, Watanabe S, Yamada T, Tsubokura Y, Nakashima H, Zhai H, Anai T, Sato S, Yamazaki T, Lü S, Wu H, Tabata S, Harada K (2012) Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc Natl Acad Sci U S A 109:E2155-2164
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316-322
Yang J, Lin R, Hoecker U, Liu B, Xu L, Wang H (2005) Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J 43:131–141
Yao D, Liu ZZ, Zhang J, Liu SY, Qu J, Guan SY, Pan LD, Wang D, Liu JW, Wang PW (2015) Analysis of quantitative trait loci for main plant traits in soybean. Genet Mol Res 14:6101–6109
Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13:2471–2481
Yue L, Li X, Fang C, Chen L, Yang H, Yang J, Chen Z, Nan H, Chen L, Zhang Y, Li H, Hou X, Dong Z, Weller JL, Abe J, Liu B, Kong F (2021) FT5a interferes with the Dt1-AP1 feedback loop to control flowering time and shoot determinacy in soybean. J Integr Plant Biol. https://doi.org/10.1111/jipb.13070
Zhang H, Nocker SV (2002) The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J 31:663–673
Zhang YM, Mao Y, Xie C, Smith H, Luo L, Xu S (2005) Mapping quantitative trait loci using naturally occurring genetic variance among commercial inbred lines of maize (Zea mays L.). Genetics 169:2267–2275
Zhang J, Song Q, Cregan PB, Nelson RL, Wang X, Wu J, Jiang GL (2015) Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genomics 16:217
Zhang J, Feng JY, Ni YL, Wen YJ, Niu Y, Tamba CL, Yue C, Song Q, Zhang YM (2017) pLARmEB: integration of least angle regression with empirical Bayes for multi-locus genome-wide association studies. Heredity 118:517–524
Zhang YM, Jia Z, Dunwell JM (2019) Editorial: the applications of new multi-locus GWAS methodologies in the genetic dissection of complex traits. Front Plant Sci 10:100
Zhang YW, Tamba CL, Wen YJ, Li P, Ren WL, Ni YL, Gao J, Zhang YM (2020) mrMLM v4.0: an R platform for multi-locus genome-wide association studies. Genom Proteom Bioinf. https://doi.org/10.1016/j.gpb.2020.06.006
Zhao X, Cao D, Huang Z, Wang J, Lu S, Xu Y, Liu B, Kong F, Yuan X (2015) Dual functions of GmTOE4a in the regulation of photoperiod-mediated flowering and plant morphology in soybean. Plant Mol Biol 88:343–355
Zheng M, Peng C, Liu H, Tang M, Yang H, Li X, Liu J, Sun X, Wang X, Xu J, Hua W, Wang H (2017) Genome-wide association study reveals candidate genes for control of plant height, branch initiation height and branch number in rapeseed (Brassica napus L.). Front Plant Sci 8:1246
Zhou L, Wang SB, Jian J, Geng QC, Wen J, Song Q, Wu Z, Li GJ, Liu YQ, Dunwell JM, Zhang J, Feng JY, Niu Y, Zhang L, Ren WL, Zhang YM (2015) Identification of domestication-related loci associated with flowering time and seed size in soybean with the RAD-seq genotyping method. Sci Rep 5:9350
Zhu G, Wang S, Huang Z, Zhang S, Liao Q, Zhang C, Lin T, Qin M, Peng M, Yang C, Cao X, Han X, Wang X, Knaap E, Zhang Z, Cui X, Klee H, Fernie AR, Luo J, Huang S (2018) Rewiring of the fruit metabolome in tomato breeding. Cell 172:249–261
Zuo Q, Hou J, Zhou B, Wen Z, Zhang S, Gai J, Xing H (2013) Identification of QTLs for growth period traits in soybean using association analysis and linkage mapping. Plant Breed 132:317–323
Acknowledgements
We thank Mr. Hanwen Zhang (hywenzhang@henceedu.com) at Hence Education Ltd., Vancouver, Canada, for improving the language within the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (32070557 and 31871242), and Huazhong Agricultural University Scientific & Technological Self-Innovation Foundation (2014RC020).
Author information
Authors and Affiliations
Contributions
YMZ designed this experiment. XH, ZRX, LZ, and CYH performed field experiments and real data analysis. XH wrote the draft, and YMZ and XH revised the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Ethical standards were not involved in the study.
Consent for publication
The authors consented for publication.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1.
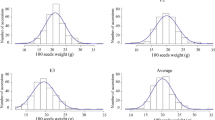
Frequency distributions for first (a) and full (b) flowering time and plant height (c) in 286 soybean accessions. NJ: Nanjing; WH: Wuhan; EZ: Ezhou. 2011 in NJ2011 indicates year. Mean ± standard deviation is showed. (PDF 2205 KB)
Supplementary Figure 2.
Validation of QTNs for first flowering time using phenotypic differences between elite and alternative alleles of each QTN. These QTNs include MM7502, MM15510, MM19192, MM25031, MM25114, MM25599, MM28606, MM30687, MM33259, MM46206, MM47999, MM48293, MM51539, MM51663, MM53568, MM53674, MM54137, MM55040, MM56754, MM57775, MM57851, MM58369, MM65212, MM67058, MM82865, MM85086, MM87394, MM92726, MM92745, MM93630, and MM102840. Error bar represents standard deviation. E1: Nanjing (2011); E2: Nanjing (2012); E5: Ezhou (2017); E6: Wuhan (2017). (PDF 525 KB)
Supplementary Figure 3.
Validation of QTNs for full flowering time using phenotypic differences between elite and alternative alleles of each QTN. These QTNs include MM5544, MM7502, MM9632, MM12021, MM14978, MM16560, MM19352, MM19374, MM20918, MM25114, MM25129, MM27076, MM32189, MM33259, MM35045, MM48293, MM51539, MM51663, MM53674, MM54137, MM55040, MM56090, MM57775, MM62301, MM62964, MM65212, MM67680, MM71416, MM72971, MM82865, MM86837, MM87394, MM92726, MM92745, MM93630, and MM99199. Error bar represents standard deviation. E1: Nanjing (2011); E2: Nanjing (2012); E5: Ezhou (2017); E6: Wuhan (2017). (PDF 537 KB)
Supplementary Figure 4.
Validation of QTNs for plant height using phenotypic differences between elite and alternative alleles of each QTN. These QTNs include MM93, MM10723, MM12824, MM14428, MM30756, MM37383, MM41287, MM45845, MM45847, MM54781, MM56083, MM63977, MM65389, MM66997, MM68942, MM77907, MM89741, MM89777, MM90929, MM95579, and MM95591. Error bar represents standard deviation. E3: Wuhan (2014); E4: Wuhan (2015); E5: Ezhou (2017); E6: Wuhan (2017). (PDF 423 KB)
Supplementary Figure 5.
The expressional levels (FPKM) of potential candidate genes for flowering time (a) and plant height (b) in 9 soybean tissues. (PDF 1117 KB)
Rights and permissions
About this article
Cite this article
Han, X., Xu, ZR., Zhou, L. et al. Identification of QTNs and their candidate genes for flowering time and plant height in soybean using multi-locus genome-wide association studies. Mol Breeding 41, 39 (2021). https://doi.org/10.1007/s11032-021-01230-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-021-01230-3