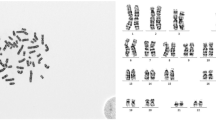
Abstract
The analysis of chromosomes is a significant and challenging task for clinical diagnosis and biological research. The technique based on color imaging is a multiplex fluorescent in situ hybridization (M-FISH), which was implemented to ease the exploration of the chromosomes. Thus, in this paper, we propose a novel quasi-Newton-based K-means clustering for the M-FISH image segmentation. Then, we use the expectation–maximization-based hierarchical Bayes model to characterize the M-FISH images. The contextual-based classification and region merging of chromosomal images is made to avoid any misclassification, and we made use of AlexNet, by modifying the activation functions of the sigmoid and softmax layer and for the optimum classification between the autosomal chromosomes and the sex chromosome. Finally, we conducted a performance analysis by measuring accuracy, recall, sensitivity, specificity, PPV, NPV, F-score, kappa, Jaccard, and Dice coefficient and compared with other existing methods and found that our proposed methodology can achieve more percentage of accuracy (6.96%) than the state of the art methods.
Graphical abstract




Similar content being viewed by others
References
Perkel JM (2019) “Chromosomal DNA comes into focus,” ed: NATURE PUBLISHING GROUP MACMILLAN BUILDING, 4 CRINAN ST, LONDON N1 9XW, ENGLAND
Nino CL, Perez GF, Isaza N, Gutierrez MJ, Gomez JL, Nino G (2018) Characterization of sex-based DNA methylation signatures in the airways during early life. Sci Rep 8:1–10
Onozato ML, Yapp C, Richardson D, Sundaresan T, Chahal V, Lee J et al (2019) Highly multiplexed fluorescence in situ hybridization for in situ genomics. J Mol Diagn 21:390–407
Madabhushi A and Lee G (2016) “Image analysis and machine learning in digital pathology: challenges and opportunities,” ed: Elsevier
Jang W, Chae H, Kim J, Son J-O, Kim SC, Koo BK et al (2016) Identification of small marker chromosomes using microarray comparative genomic hybridization and multicolor fluorescent in situ hybridization. Mol Cytogenet 9:61
Sharma M, Jindal S, and Vig L (2020) “Method and system for automatic chromosome classification,” ed: Google Patents
Lukumbuzya M, Schmid M, Pjevac P, Daims H (2019) A multicolor fluorescence in situ hybridization approach using an extended set of fluorophores to visualize microorganisms. Front Microbiol 10:1383
Brich S, Bozzi F, Perrone F, Tamborini E, Cabras AD, Deraco M, et al (2019) “Fluorescence in situ hybridization (FISH) provides estimates of minute and interstitial BAP1, CDKN2A, and NF2 gene deletions in peritoneal mesothelioma”. Modern Pathol 1–11
Patel SH, Batchala PP, Mrachek EKS, Lopes M-BS, Schiff D, Fadul CE et al (2020) MRI and CT identify isocitrate dehydrogenase (IDH)-mutant lower-grade gliomas misclassified to 1p/19q codeletion status with fluorescence in situ hybridization. Radiology 294:160–167
Shen X, Qi Y, Ma T, Zhou Z (2019) A dicentric chromosome identification method based on clustering and watershed algorithm. Sci Rep 9:1–11
Ooi A, Inokuchi M, Horike S-I, Kawashima H, Ishikawa S, Ikeda H et al (2019) Amplicons in breast cancers analyzed by multiplex ligation-dependent probe amplification and fluorescence in situ hybridization. Hum Pathol 85:33–43
Sangpakdee W, Phimphan S, Liehr T, Fan X, Pinthong K, Patawang I et al (2016) Characterization of chromosomal rearrangements in pileated gibbon (Hylobates pileatus) using multiplex-FISH technique. The Nucleus 59:131–135
Ratan ZA, Zaman SB, Mehta V, Haidere MF, Runa NJ, and Akter N (2017) “Application of fluorescence in situ hybridization (FISH) technique for the detection of genetic aberration in medical science”. Cureus 9
Qi G, Guan W, He Z, Huang A (2019) Adaptive kernel fuzzy C-Means clustering algorithm based on cluster structure. J Intell Fuzzy Syst 37:2453–2471
Kapil S, Chawla M, and Ansari MD (2016) “On K-means data clustering algorithm with genetic algorithm,” in 2016 Fourth International Conference on Parallel, Distributed and Grid Computing (PDGC) 202–206
Zeinalkhani L, Jamaat AA, Rostami K (2018) Diagnosis of brain tumor using combination of K-means clustering and genetic algorithm. Front Health Inform 7:6
Baheti B, Ahuja G, and Parode A (2016) “Automatic classification of M-FISH human chromosome images using fuzzy classifier and statistical classifier images using fuzzy classifier and statistical classifier,” in International Conference on Communication and Signal Processing 2016 (ICCASP 2016)
Menaka D and Vaidyanathan SG (2019) “Expectation maximization segmentation algorithm for classification of human genome image,” in 2019 3rd International Conference on Computing Methodologies and Communication (ICCMC) 1055–1059
Qin Y, Wen J, Zheng H, Huang X, Yang J, Song N et al (2019) Varifocal-net: a chromosome classification approach using deep convolutional networks. IEEE Trans Med Imaging 38:2569–2581
Esfahanian P and Akhavan M (2019) “GACNN: training deep convolutional neural networks with genetic algorithm,” arXiv preprint arXiv:1909.13354
Sampat MP, Castleman K, and Bovik A (2002) “Pixel-by-pixel classification of MFISH images,” in Proceedings of the Second Joint 24th Annual Conference and the Annual Fall Meeting of the Biomedical Engineering Society][Engineering in Medicine and Biology 999–1000
Sampat MP, Bovik AC, Aggarwal JK, Castleman KR (2005) Supervised parametric and non-parametric classification of chromosome images. Pattern Recogn 38:1209–1223
Choi H, Castleman KR, and Bovik AC (2004) “Joint segmentation and classification of M-FISH chromosome images,” in The 26th Annual International Conference of the IEEE Engineering in Medicine and Biology Society 1636–1639
Wang YP, Castleman KR (2005) Normalization of multicolor fluorescence in situ hybridization (M-FISH) images for improving color karyotyping. Cytom Part A J Int Soc Anal Cytol 64:101–109
Karvelis PS, Fotiadis DI, Georgiou I, and Syrrou M (2006) “A watershed based segmentation method for multispectral chromosome images classification,” in 2006 International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 3009-3012
Schwartzkopf WC, Bovik AC, Evans BL (2005) Maximum-likelihood techniques for joint segmentation-classification of multispectral chromosome images. IEEE Trans Med Imaging 24:1593–1610
Wang Y-P and Dandpat AK (2006) “A hybrid approach of using wavelets and fuzzy clustering for classifying multispectral florescence in situ hybridization images”. Int J Biomed Imaging 2006
Wang X, Zheng B, Li S, Zhang R, Mulvihill JJ, Chen WR, et al (2009) “Automated detection and analysis of fluorescent in situ hybridization spots depicted in digital microscopic images of Pap-smear specimens”. J Biomed Optics 14:021002
Choi H, Bovik AC, Castleman KR (2008) Feature normalization via expectation maximization and unsupervised nonparametric classification for M-FISH chromosome images. IEEE Trans Med Imaging 27:1107–1119
Karvelis PS, Likas AC (2013) Fully unsupervised m-FISH chromosome image characterization. IEEE J Biomed Health Inform 17:1068–1078
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kanimozhi, V.S., Balasubramani, M. & Anuradha, R. Hierarchal Bayes model with AlexNet for characterization of M-FISH chromosome images. Med Biol Eng Comput 59, 1529–1544 (2021). https://doi.org/10.1007/s11517-021-02384-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-021-02384-0