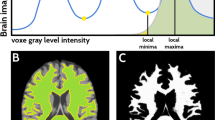
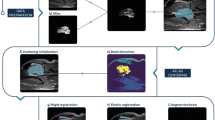
Abstract
Neurologist analyses shape and structure of brain parts through any medical images such as CT, MRI, and PET for disease diagnosis. For diagnosis, automatic medical image segmentation segments the parts of brain with low contrast, and artefacts are never removed over boundary region in different parts of brain. Manual segmentation shows poor differentiation in boundary regions due to artefacts or steaks. In this paper, we propose dyadic CAT optimisation (DCO) algorithm for segmenting the brain regions from CT and MRI images via nonlinear perspective foreground and background projection. DCO algorithm provides exact structure and shape of brain regions and eliminates artefacts in boundary regions. DCO algorithm delineates the boundary region such as dentate nucleus, pontine tegmentum, pontine nuclei, petrosal nerve, petrous part of temporal bone, crista galli, internal occipital crest, and mastoid emissary foramen in brain image with high visibility and enhanced boundary and differentiates deformable shape. Performance of DCO algorithm is evaluated through 50 MRI and CT brain images and eight images with complex bone and muscle mass structures of brain. DCO algorithm shows an accuracy of 90% through structural similarity index.











Similar content being viewed by others
Code availability
The code is available from the corresponding author on reasonable request.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Tian, J., Member, S., Xue, J., Dai, Y., Chen, J., Zheng, J.: A novel software platform for medical image processing and analyzing. IEEE Trans. Inf. Technol. Biomed. 12(6), 800–812 (2008)
Von Landesberger, T., Basgier, D., Becker, M.: Comparative local quality assessment of 3D medical image segmentations with focus on statistical shape model-based algorithms. IEEE Trans. Vis. Comput. Graph. 22(12), 2537–2549 (2015)
Chittajallu, D.R., Paragios, N., Kakadiaris, I.A.: An explicit shape-constrained MRF-based contour evolution method for 2-D medical image segmentation. IEEE J. Biomed. Heal. Inform. 18(1), 120–129 (2014)
Franchini, S., et al.: ConformalALU: a conformal geometric algebra coprocessor for medical image processing. IEEE Trans. Comput. 64(4), 955–970 (2015)
Qi, G.U.O., Long, W., Shuting, S.: Multiple-channel local binary fitting model for medical image segmentation∗. Chin. J. Electron. 24(4), 56 (2015)
Khadidos, A., Sanchez, V., Li, C.: Weighted level set evolution based on local edge features for medical image segmentation. IEEE Trans. Image Process. 26(4), 1979–1991 (2017)
Doshi, T., et al.: Combining interpolation and 3D level set method ( I + 3DLSM ) for medical image segmentation. Electron. Lett. 52(8), 592–594 (2016)
Pratondo, A., Chui, C., Ong, S.: Robust edge-stop functions for edge-based active contour models in medical image segmentation. IEEE Signal Process. Lett. 23(2), 222–226 (2016)
Karasev, P., Kolesov, I., Fritscher, K., Vela, P., Mitchell, P., Tannenbaum, A.: Interactive medical image segmentation using PDE control of active contours. IEEE Trans. Med. Imaging 32(11), 2127–2139 (2013)
Ellis, H., Logan, B.M., Dixon, A.K., Bowden, D.J.: Atlas of Body Sections, CT and MRI Images (2015)
Funding
No funding.
Author information
Authors and Affiliations
Contributions
Conceptualization, Raja Paul Perinbam; Methodology, R. Partheepan, Raja Paul Perinbam. M. Krishnamurthy; Formal Analysis, R. Partheepan, M. Krishnamurthy and N. R. Shanker; Resources, N.R. Shanker; Writing—Original Draft Preparation, R. Partheepan, Raja Paul Perinbam; Writing—Review and Editing, N.R. Shanker; Supervision, Raja Paul Perinbam.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
All procedures were in accordance with the 1964 Helsinki Declaration (and its amendments). No approval by ethical committee or institutional review board was required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Partheepan, R., Raja Paul Perinbam, J., Krishnamurthy, M. et al. Visualization of dentate nucleus, pontine tegmentum, pontine nuclei from CT image via nonlinear perspective projection. SIViP 16, 137–145 (2022). https://doi.org/10.1007/s11760-021-01973-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-021-01973-8