Abstract
Rhodococcus spp. strains are widespread in diverse natural and anthropized environments thanks to their high metabolic versatility, biodegradation activities, and unique adaptation capacities to several stress conditions such as the presence of toxic compounds and environmental fluctuations. Additionally, the capability of Rhodococcus spp. strains to produce high value-added products has received considerable attention, mostly in relation to lipid accumulation. In relation with this, several works carried out omic studies and genome comparative analyses to investigate the genetic and genomic basis of these anabolic capacities, frequently in association with the bioconversion of renewable resources and low-cost substrates into triacylglycerols. This review is focused on these omic analyses and the genetic and metabolic approaches used to improve the biosynthetic and bioconversion performance of Rhodococcus. In particular, this review summarizes the works that applied heterologous expression of specific genes and adaptive laboratory evolution approaches to manipulate anabolic performance. Furthermore, recent molecular toolkits for targeted genome editing as well as genome-based metabolic models are described here as novel and promising strategies for genome-scaled rational design of Rhodococcus cells for efficient biosynthetic processes application.
Similar content being viewed by others
Introduction
Rhodococcus genus comprises Gram-positive, non-motile, non-sporulating, aerobic bacteria, with a high G + C content and a mycolic acid-containing cell wall (Martínková et al. 2008). Members of Rhodoccocus genus are widely distributed in soil, water, and marine sediments, due to their metabolic flexibility and their tolerance to various stresses (e.g., presence of toxic metals and metalloids, desiccation, low nutrient availability, high concentration of organic pollutants). Only a few strains are pathogens of plants and animals (including humans) and belong to Rhodococcus fascians and Rhodococcus equi species, respectively (Cappelletti et al. 2019). The outstanding metabolic diversity and strong persistence of this genus is associated to peculiar features of their cell surface (i.e., hydrophobicity, mycolic acids content, and fatty acid composition rearrangement) and, in some cases, to the presence of large and complex genomes containing a multiplicity of genes involved in unique catabolic and anabolic processes (De Carvalho et al. 2014; Laczi et al. 2015; Orro et al. 2015; Cappelletti et al. 2016; Kis et al. 2017; Presentato et al. 2016, 2018a, b, c, 2020). In relation to this, Rhodococcus spp. strains are capable of performing biotransformation and biodegradation of many organic and xenobiotic compounds, such as hydrocarbons and chlorinated hydrocarbons, naphthenic acids, nitroaromatics, and pharmaceuticals (e.g., diclofenac and sulfamethoxazole) (Auffret et al. 2009; Cappelletti et al. 2010, 2015, 2016, 2018; Orro et al. 2015; Presentato et al. 2018a, b, c; Weidhass et al. 2009; Ivshina et al. 2019; Tyumina et al. 2019; Larcher and Yargeau 2011). Due to these wide metabolic capabilities and stress resistance/tolerance, Rhodococcus strains are considered ideal candidates for biotechnological applications in environmental remediation and pharmaceutical and chemical industries (Bell et al. 1998; Busch et al. 2019; Ceniceros et al. 2017; Kis et al. 2015; Larkin et al. 2005; van der Geize and Dijkhuizen 2004, Patek et al. 2021). In particular, extensive research focused on the production and accumulation of neutral lipids, mostly triacylglycerols (TAGs), in Rhodococcus through the bioconversion of industrial wastes and lignin biomass feedstocks. TAGs are energy-rich compounds formed by a molecule of glycerol esterified with three fatty acids chains. They can be used for biofuel generation after bacterial cell extraction through conventional chemical and physical extraction methods (e.g., sonication, solvent extraction) or through biological approaches (e.g., enzymatic cell lysis, phage-based extraction) (Hwangbo and Chu 2020).
Other valuable compounds that can be produced by Rhodococcus spp. strains are glycolipid biosurfactants, carotenoids, polyhydroxyalkanoates (PHAs), metal-based nanomaterials, and novel antimicrobials (Cappelletti et al. 2020). While experimental works have mostly investigated culture conditions and suitable substrates for efficient biotechnological applications, the identification of specific genes/proteins involved in these biosynthetic pathways has been for a long time hindered by the lack of efficient molecular methods/tools generally applicable to Rhodococcus spp. strains (Cappelletti et al. 2010). Transformation protocols in Rhodococcus spp. strains initially relied on protoplast-mediated procedures (Singer and Finnerty 1988; Duran 1998; Desomer et al. 1990). More recently, conjugation and electroporation methods have been successfully applied, although the results were shown to greatly vary within the Rhodococcus genus at species and even strain level (Shao et al. 1995; Sekizaki et al. 1998; Kalscheuer et al. 1999).
The decreasing cost of sequencing technologies and increasing number of complete genome sequences of different Rhodococcus spp. strains available in the databases allowed the in silico detection of numerous genes/enzymes involved in biosynthetic processes mostly leading to the accumulation of lipids. In this regard, genes predicted to be involved in the synthesis and accumulation of triacylglycerols, wax esters, polyhydroxyalkanoates (PHAs), and fatty acids were described in R. jostii RHA1, R. opacus PD630, R. opacus B4, R. erythropolis PR4, R. equi 103S, and R. fascians F7 (Hernandez et al. 2008; Alvarez et al. 2013; Cappelletti et al. 2019).
Among the different Rhodococcus strains that have been described in the literature, R. opacus PD630 and R. jostii RHA1 have been the most extensively studied for their biosynthetic and bioconversion activities due to the following reasons, (i) the peculiar capacity of using lignocellulose-based sugars along with toxic lignin-derived aromatic compounds; (ii) the high content of valuable lipids, mostly triacylglycerols (TAGs), that can accumulate from different carbon sources; (iii) the rapid growth rate; and finally, (iv) a good genetic tractability (DeLorenzo et al. 2017). Indeed, under nitrogen-limited conditions (i.e., reduced amount of nitrogen source added to the growth medium) and optimized fed-batch fermentation conditions, an accumulation of TAGs corresponding to around 75% of the cell dry mass can be achieved in R. opacus PD630 cells growing on glucose (Kim et al. 2019). On the other hand, some genetic manipulation strategies were developed and successfully applied to R. opacus PD630 and R. jostii RHA1 that allowed both genetic analyses and strain performance improvement.
Previous reviews summarize the regulation and the metabolic pathways involved in lipid accumulation in Rhodococcus (Alvarez et al. 2019), the different valuable compounds biosynthesized by members of this genus (Cappelletti et al. 2020) and those specifically derived from lignocellulose/lignin bioconversion (Anthony et al. 2019). Conversely, the present review is focused on the main genetic manipulation and metabolic engineering strategies applied to modify this genus strains to extend and optimize their biosynthetic capabilities and biotechnological applications (summarized in Table 1; Figs. 1 and 2). Some indications on novel genome editing tools (i.e., based on CRISPR/Cas9 and recombineering systems) are also reported as new strategies for rational genetic engineering of Rhodococcus spp. strains for biosynthetic purposes.
Main pathways in Rhodococcus spp. strains leading to the production and accumulation of valuable compounds from lignin degradation products (A) and steroid (B). The colors of the arrows indicate the methodological approaches used to optimize/improve the specific biosynthetic capability, i.e., red and green arrows correspond to heterologous expression and genome-based metabolic engineering, respectively. Dashed arrows represent several reactions, while solid arrows indicate a single reaction
Adaptive laboratory evolution
Regarding biosynthetic and bioconversion strategies, several adaptive laboratory evolution (hereafter: ALE) approaches were applied to generate a set of R. opacus PD630 strains featured by optimized growth and TAG production and accumulation from substrates derived from lignocellulosic biomass (Kurosawa et al. 2015a, b; Yoneda et al. 2016; Henson et al. 2018). ALE experiments consisted in the construction of mutant populations starting from Rhodococcus spp. strain cultures which were sequentially transferred into new cultures, at regular intervals, in the presence of specific growth conditions and/or stressors (i.e., selective pressure) (Fig. 1). This re-inoculation procedure carried out several times promotes the selection and propagation of beneficial mutations that sustain improved growth rate and/or higher resistance/tolerance capacities (i.e., improved fitness) (Dragosits and Mattanovich 2013; Sandberg et al. 2019).
In particular, to expand growth substrates spectrum, an ALE approach was applied to improve glycerol utilization in the engineered xylose-fermenting R. opacus strain MITXM-61, originating from R. opacus PD630 (Kurosawa et al. 2014, 2015b). In this work, sequential transfers of liquid cultures were performed by supplying at each passage 100 g/L of glycerol as sole carbon source. This procedure allowed the isolation of the strain MITGM-173, which showed improved growth performance in the presence of glycerol as compared to the parental strain. Strain MITGM-173 also exhibited the ability to simultaneously metabolize glucose, xylose, and glycerol to produce a large number of TAGs. TAG production resulted to be higher when the glycerol was added after 2 days of growth on glucose and xylose. Although glycerol assimilation mechanism needs further investigation, the authors hypothesized that the improved lipid production could be due to the function of glycerol as direct precursor in TAG biosynthesis (Kurosawa et al. 2015b). In another work by the same authors, ALE experiments were performed to generate a Rhodococcus strain with enhanced tolerance to the inhibitors derived from lignocellulose hydrolysis, such as furans and phenols (Kurosawa et al. 2015a). Specifically, a R. opacus MITXM-61 evolved strain was obtained that was tolerant to lignin, 4-HB, and syringaldehyde. The mutant also showed improved growth performance and a higher amount of TAG production on lignocellulosic hydrolysates as compared to the parental strain.
More recent studies combined ALE with multi-omics approaches to identify novel targets for engineering R. opacus strains toward lignin valorization. Specifically, in two different papers, ALE was successfully applied to PD630 obtaining evolved strains with improved aromatic utilization and tolerance and lipid production (Yoneda et al. 2016; Henson et al. 2018). Comparative transcriptomics studies between the WT and the adaptively evolved strains showed that both strains upregulate gene clusters involved in the transformation of lignin-derived aromatic compounds to either catechol (CAT) or protocatechuate (PCA) which are subsequently converted through the β-ketoadipate pathway in succinyl-CoA and acetyl-CoA. Acetyl-CoA is the main precursor of lipid biosynthesis. Interestingly, in all adaptively evolved strains, the 36% of non-synonymous SNPs were identified in genes involved in redox reactions, suggesting that the redox state of the cell could be important for the improved tolerance and utilization of aromatic compounds. In particular, all mutant strains shared a non-synonymous SNP in the gene encoding for the superoxide dismutase (SOD), causing a 56% activity loss under in-vitro conditions (Henson et al. 2018). A lower SOD activity could increase the intracellular levels of superoxide radicals that participate in the oxidation of aromatic compounds catalyzed by mono- and dioxygenases (Gatti et al. 1994; Bugg 2001; Henson et al. 2018). An additional metabolomic study elucidated the biodegradation pathway of a lignin-derived aromatic compound, i.e., phenol, via quantitative flux balance analysis (FBA). In the presence of phenol, PD630 metabolic network showed strong metabolic fluxes through the TCA cycle, making this strain an ideal host for the production of metabolic intermediates such as acetyl-CoA and α-ketoglutarate for TAG production from lignocellulosic biomass (Roell et al. 2019).
Heterologous gene expression
The production of specific proteins and functions in Rhodococcus spp. strains was achieved by developing several types of expression vectors based on cryptic (i.e., with unknown function) plasmids and transposable DNA elements tools for the insertion of cloned genes into chromosomes (Mitani et al. 2006). In the field of bioconversions, Rhodococcus strains were engineered to improve the utilization of cellulose and hemicellulose degradation products such as cellobiose, xylose, l-arabinose, and levoglucosan (Fig. 2A). The latter is an anhydrous sugar deriving from the pyrolysis of cellulose, and only few microorganisms are known to be able to metabolize it. With regard to the recombinant strains, R. opacus PD630 engineered with the bglABC operon from Thermobifida fusca, which encodes for two ABC sugar transport proteins (BglA and BglB) and a β-glucosidase (BglC). As a result, the recombinant strain exhibited both increased growth on cellobiose and improved lipid accumulation (up to 39% of the cell dry mass) as compared to the wild-type strain (Hetzler and Steinbüchel 2013).
Heterologous xylA and xylB genes, coding for d-xylose isomerase and xylulokinase respectively, from Streptomyces padanus MITKK-103 (Kurosawa et al. 2013) and from Streptomyces lividans TK23 (Xiong et al. 2012) conferred to R. opacus PD630 and R. jostii RHA1 the ability to produce lipids in presence of xylose as sole carbon source. The two works used different approaches to achieve the heterologous expression of xyl genes. Kurosawa et al. (2013) created a genomic library of S. padanus in R. opacus PD630 cells that were further screened for their ability to grow on xylose. Within this library, the strain Xsp8 showed the highest level of TAG accumulation. Characterization of the Xsp8 plasmid confirmed the presence of S. padanus genes homologous to xylA and xylB. The recombinant strain was able to grow simultaneously on glucose and xylose with final TAG accumulation corresponding to 45.8% of the cell dry mass (CDM). On the other hand, Xiong et al. (2012) cloned xylA and xylB genes from S. lividans inside the E. coli-Rhodococcus shuttle vector pNV18 downstream of the inducible tac promoter (Ptac). Under nitrogen-limited condition, the lipid accumulation on xylose resulted to be up to 68.3% and 52.5% of CDM in PD630 and RHA1 recombinant cells, respectively (Xiong et al. 2012). These Rhodococcus strains were also engineered to obtain recombinant cells able to use l-arabinose for the growth. The genes araB, araD, and araA from Streptomyces cattleya NRRL 8057 (Kurosawa et al. 2015c) and from Escherichia coli K12 MG1655 (Xiong et al. 2016a) were expressed in R. opacus PD630 and R. jostii RHA1, respectively. The three genes code for the enzymes l-ribulokinase, l-ribulose-5-phosphate 4-epimerase, and l-arabinose isomerase which catalyze the reactions leading to l-arabinose transformation into d-xylulose-5-phosphate that can enter the pentose phosphate pathway. The engineered PD630 strain MITAE-348 exhibited good growth performance in the presence of high concentrations of l-arabinose (up to 100 g/L) and subsequent lipid accumulation (39.7% of the CDM). In the presence of a mixture of l-arabinose and d-glucose (1:1), the strain metabolized both sugars simultaneously with higher TAG production as compared to growth on the only l-arabinose, reaching a TAG content that corresponds to 42.0% of the CDM (Kurosawa et al. 2015c). In addition to the capacity to metabolize l-arabinose, recombinant cells of R. jostii RHA1 showed an increase in biomass production when the arabinose transporter operon araFGH was expressed, while higher lipid content (56.8% of the CDM) was obtained by expressing atf1 gene, a key gene for TAG biosynthesis, from R. opacus PD630 (Xiong et al. 2016a). The heterologous expression approach was also successfully applied to R. jostii RHA1, conferring the capacity to metabolize the sugar levoglucosan by expressing the gene levoglucosan kinase (lgk) from the yeast Lipomyces starkeyi YZ-215. The gene lgk encodes a specific levoglucosan kinase that converts levoglucosan into glucose-6-phosphate, which enters the glycolysis pathway. As a result, the recombinant RHA1 cells acquired the capacity to grow on levoglucosan as sole carbon source. Despite the lipid accumulation rate on this substrate was lower, the final lipid content on levoglucosan resulted to be similar to that obtained on glucose, up to 43.54% of the cell dry mass (Xiong et al. 2016b).
Overexpression of autologous genes
Overexpression of native genes in Rhodococcus spp. strains associated with the improvement of biosynthetic abilities was mainly performed by cloning these genes in episomal expression systems under the control of inducible promoters (e.g., PtipA and Pace inducible by thiostrepton and acetamide, respectively). The fatty acid biosynthesis was enhanced in R. opacus PD630 grown on glucose as sole carbon source by overexpression of autologous thioesterases (Huang et al. 2016). Thioesterases (TEs) are hydrolytic enzymes that break the thioester bond between acyl-ACP and fatty acid chain stopping the fatty acid elongation cycle. Released fatty acids can be converted to TAGs and other storage lipids. The authors found 4 putative TE genes in R. opacus PD630. Overexpression of TE2 and TE4 led to an increased lipid content reaching 46% and 44% of cell dry mass (CDM) respectively, whereas the control strain and the recombinant cells overexpressing TE1 and TE3 only reached 37% of CDM. Overexpression and deletion of additional genes involved in TAG metabolism, such as atf1 and atf2, pap2, tadD, ltp1, and nlpR, and genes encoding a NADP+-dependent malic enzyme were conducted in order to clarify their functional role in the lipid metabolism in R. opacus PD630 and R. jostii RHA1 (Hernández et al. 2013, 2015, 2017, 2019; Mandal et al. 2019; MacEachran and Sinskey 2013). Although these works were not focused on the evaluation of metabolic and bioconversion improvement, the overexpression or deletion of the aforementioned genes led to changes in TAG accumulation and yield, suggesting a possible application in genetic and metabolic engineering.
Genome-scale metabolic models and metabolic engineering
The representation of biological systems complexity by mathematical models can be used to describe and predict cell behavior and metabolism and therefore useful for designing metabolic engineering strategies for biosynthetic pathway assessment and optimization. In this respect, genome-scale metabolic models (GEMs), i.e., mathematical representations of the stoichiometry of the biochemical networks, can be used to integrate in one single model information retrieved from physiological studies, gene-protein-reaction association, metabolic flux analysis, and thermodynamic analysis of pathways (O’Brien et al. 2015). For instance, GEM can be used to redesign portions of the metabolic network and predict the production rate of a metabolite under specific operating conditions. Redesign the metabolic network can be achieved by imposing metabolic constraints and in silico by removing a given reaction from the GEM (e.g., gene knock-out mutant) or imposing uptake null for a nutrient in the growth media. In this regard, the GEM model iMT1174 was developed in R. jostii RHA1 and used to predict the accumulation rate of three types of carbon storage compounds (i.e., glycogen, polyhydroxyalkanoates, and triacylglycerols) using different carbon sources (glucose or acetate) and under growth conditions typically occurring in activated sludge bioreactor systems for wastewater recovery (Tajparast and Frigon 2015, 2018). Starting from the iMT1174 metabolic network, different objective functions were implemented based on a set of metabolic constraints such as minimization and maximization of the specific metabolic fluxes and minimization and maximization of ATP production rate and reducing redox potential (NADH). The predicted accumulation rate of storage compounds for each of the objective functions was further validated using 13C-metabolic flux analysis (13C-MFA) (Tajparast and Frigon 2015). These studies, taken together, represent the first effort of simulating and predicting Rhodococcus metabolic fluxes leading to the production and accumulation of valuable compounds with industrial interest.
A few metabolic engineering approaches were focused on the genetic and metabolic modification of R. opacus PD630 and R. jostii RHA1 to promote lignin bioconversion and enhance the production of valuable compounds. These works focused on the optimization of the bioconversion of lignin into TAGs (Xie et al. 2019), cis,cis-muconate (CCMA) (Cai et al. 2020), or pyridine-dicarboxylic acids (Spence et al. 2021) and are summarized in Fig. 2A. In particular, the production of TAGs from lignin in R. opacus PD630 was improved by optimizing (at both transcriptional and translation level) the heterologous expression of a laccase gene (from Streptomyces coelicolor) and the secretion system (i.e., Tat transporter components, TatA and TatC) that is needed for the laccase extracellular ligninolytic activity (Xie et al. 2019). Furthermore, to allocate more carbon into lipid biosynthesis, the type I fatty acid synthase (fasI) operon encoding the main enzyme involved in fatty acid biosynthesis in Rhodococcus (Schweizer and Hofmann 2004) and the diacylglycerol acyltransferase gene atf2, which catalyzes the final step of TAG production (Hernández et al. 2013) were overexpressed in the laccase-producing PD630 strain. As a result, the final recombinant strain showed tenfold increased growth on 1% w/v of insoluble kraft lignin as compared to the wild-type strain and exhibited a significantly enhanced lipid production (Xie et al. 2019). With regard to the bioconversion of lignin into cis,cis-muconate (CCMA), Cai et al. (2020) genetically modified R. opacus PD630 by introducing two genes from Enterobacter cloacae, which encode a protocatechuate decarboxylase and a phenyltransferase. The introduction of these enzymes was aimed at funneling lignin degradation intermediates (i.e., protocatechuate) to the catechol degradation pathway that leads to the production of CCMA through β-ketoadipate pathway. Genes involved in further CCMA degradation and in alternative degradation pathways of catechol and protocatechuate were deleted to optimize the CCMA accumulation from lignin and lignin-derived aromatics. Both deletion and insertion were obtained by applying a markerless gene deletion/insertion system on a R. opacus PD630 mutated in a phenylalanyl-tRNA synthase gene that is used as negative counter-selection marker.
Recently, lignin was also found to be converted in pyridine-dicarboxylic acids (PDCAs) by an engineered R. jostii RHA1, for possible new bioplastics development (Spence et al. 2021). In this work, a metabolic engineering approach was applied to RHA1 to re-route the protocatechuate (derived from lignin and lignin-derived compounds) into the production of PDCA. In particular, ligAB genes from Sphingobium SYK-6 or praA gene from Paenibacillus sp. JJ-1b were inserted in the place of pcaHG genes in the RHA1 chromosome by homologous recombination. This method simultaneously allowed to block the competitive β-ketoadipate pathway by deleting the gene coding for the first enzyme of the pathway (pcaHG) and to express the enzymes responsible for the production of pyridine-2,4-dicarboxylic acid (2,4-PDCA) and pyridine-2,5-dicarboxylic acid (2,5-PDCA), respectively. The additional heterologous expression of dyp2 gene from Amycolatopsis sp. 75iv2, coding for a peroxidase in the mutant strain, significantly increased the rate of lignin oxidation and accumulation of 2,4-PDCA in the final recombinant RHA1 strain utilizing different types of lignin (Spence et al. 2021).
While these works were mainly focused on valuable compound production from lignin bioconversion, a study by Kim et al. (2019) described the optimization of Rhodococcus biosynthetic abilities with regard to the high value-added products (i.e., free fatty acids) deriving from TAGs biotransformation (Fig. 2A). With this aim, the genome scale metabolic modeling was combined with a systematic metabolic engineering strategy in order to predict R. opacus PD630 metabolism and to estimate the theoretical yields of fatty acids and their derivatives. In particular, the cultivation conditions of R. opacus were optimized to maximize the accumulation of TAGs from glucose metabolism and, after this, the metabolism of the strain was systematically analyzed and redesigned to enable higher production yield of fatty acids and biodiesel (i.e., fatty acid ethyl esters and long-chain hydrocarbons). The authors performed the deletion of specific genes to block intermediate or products degradative pathways, replacement of some native promoters to enhance the lipid production, and insertion of additional genes to introduce heterologous metabolic pathways involved in fatty acid ethyl ester accumulation. As a result, three recombinant strains were obtained which showed the highest ability ever described through fermentative processes to produce free fatty acids (FFAs, up to 50.2 g/L), fatty acid ethyl esters (FAEEs, up to 21.3 g/L), and long-chain hydrocarbons (LCHCs up to 5.2 g/L) (Kim et al. 2019).
An additional study by Guevara et al. (2019) described the genetic modification of Rhodococcus ruber Chol-4 to produce and accumulate testosterone from 4-androsterone-3,17-dione (AD) bioconversion (Fig. 2B). Rhodococcus ruber Chol-4 is well known as a steroid degrader, although it is not a model strain and therefore not extensively characterized in the literature. For this reason, unlike Rhodococcus strains PD630 and RHA1 for which molecular tools for genetic manipulation are well known and efficient, a novel method for gene deletion (based on pK18-derived plasmid) was developed to manipulate Chol-4 strain. A specific expression vector was also constructed for this strain by modifying the Nocardia-E. coli shuttle vector pNV119 harboring the inducible promoter PnitA. Using these methods, a Chol-4 strain deletion mutant was developed to prevent AD and testosterone degradation by blocking the steroid catabolic pathway. Then, the gene coding for the enzyme 17-ketosteroid reductase (17ß-HSD) from the fungus Cochliobolus lunatus was inserted to allow the biotransformation of AD into testosterone. The recombinant strain showed a molar conversion rate of AD to testosterone after 24 h of 48.2%, while no production was detected in the wild-type strain. When glucose was added to the growth medium as co-substrate to stimulate the nicotinamide cofactor regeneration, which is involved in the reaction, the molar conversion rate of AD to testosterone after 24 h increased to the 61.5% (Guevara et al. 2019).
First synthetic biology approaches applied to Rhodococcus spp. strains
Recent advances in genomics and genome editing opened new perspectives for the engineering of Rhodococcus spp. strains based on genomic-scale rational design and synthetic biology approaches. In this context, specific genetic tools for tunable gene expression were characterized and developed to expand the ability to control and characterize gene expression in Rhodococcus spp. strains. Indeed, the construction of synthetic biology circuits and pathways relies on well-defined libraries of promoter components (i.e., ribosome-binding site and regulatory regions) that in a controlled and predictable way can drive the expression of target genes. Promoter mini-pools with different activity levels were developed in R. opacus PD630 and R. ruber TH by using different fluorescent reporter genes, β-galactosidase (LacZ), and nitrilase (NHase) as different promoter activity probes (Jiao et al. 2018; DeLorenzo et al. 2017). In detail, the three chemically inducible promoters pBAD, pTet, and pAcet deriving from E. coli or Mycobacterium smegmatis were optimized for R. opacus PD630 as well as two classes of metabolite sensors responsive to nitrogen levels and specific aromatic monomers, typically found in depolymerized lignin, e.g., phenol, 4-hydroxybenzoic acid, guaiacol.
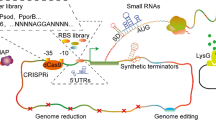
Although promoter libraries, reporter genes, and shuttle vectors have been developed for Rhodococcus as genetic tools, the genome editing of these bacteria has been hampered by its high GC content (~ 70%) and by low transformation/recombination efficiency (DeLorenzo et al. 2017, 2018; Jiao et al. 2018; Liang et al. 2020). Recently, DeLorenzo et al. (2018) developed a recombineering method for site-specific gene insertion and deletion, which was based on the activity of the bacteriophage recombinases Che9c60 and Che9c61 in PD630. This study provided the groundwork to develop a CRISPR/Cas9-mediated triple-plasmid recombineering system for genetic engineering of R. ruber TH. Specifically, the stable mutant strain R. ruber THY was obtained by using this CRISPR/Cas9-based method which featured an increased acrylamide production capacity as a result of the nitrile hydratase point mutation and a by-product gene deletion. Unlike R. opacus strains, the by-pass of restriction-modification system seemed to be necessary for efficient transformation of the R. ruber strains (Liang et al. 2020). Additional CRISPR-based approaches concerned the utilization of a CRISPR interference (CRISPRi) and a codon-optimized version of the catalytically dead Cas9 (dCas9) deriving from Streptococcus thermophilus (dCas9Sth1), as a system for gene expression control in R. opacus PD630. Like in Mycobacterium tuberculosis, the repression ability of dCas9Sth1 was independent of the distance of the sgRNA (single-guide RNA) from the transcriptional start site (DeLorenzo et al. 2018). In a very recent study, the possibility to utilize genetic parts to build genetic circuits to perform “logic function” in R. opacus was also explored. Genetic logic circuits—AND, NAND, and IMPLY—were constructed in PD630 strain combining a T7 RNA polymerase-based expression system (T7 RNAP), three novel synthetic IPTG-dependent promoters, and four aromatic sensors (DeLorenzo and Moon 2019). Apparently, while these recent studies provide the groundwork of the application of synthetic biology strategies to R. opacus PD630, further work is required to genetically characterize other Rhodococcus species which are known to be highly diversified in terms of genomic contents, metabolic pathways, and evolutionary adaptations and therefore possibly usable for different biotechnological applications (Cappelletti et al. 2019).
Conclusions
Members of Rhodococcus genus are able to use low-cost and renewable resources as bioconversion substrates for the production of high value-added compounds. In this field, Rhodococcus bioconversion of lignocellulosic biomass into neutral lipids, i.e., triacylglycerols (TAGs) for biofuel generation, represents the most prominent example. Genetic manipulation strategies based on approaches of adaptive laboratory evolution (ALE) and expression systems of specific autologous or heterologous genes have successfully led to the generation of Rhodococcus spp. strains with improved biosynthetic activities in terms of production yields, types and number of metabolized substrates, tolerance/resistance to lignin-derivative stressors (Table 1). The mutagenesis experiments were in some cases combined with multi-omic approaches and genome-based metabolic modeling to provide integrative and system-level information about metabolic pathways involved in the biosynthetic process in R. jostii RHA1 and R. opacus PD630 and genetic traits to be targeted in possible genome editing approaches. Interestingly, a few metabolic engineering approaches were successfully applied to R. opacus PD630 strains (by deleting genes that catabolize the desired product or metabolic intermediate and by introducing new functions), which led to the development of strains characterized by outstanding capacities, i.e., the highest efficiency in producing fatty acids and related products ever reported. Recent breakthroughs in genetic engineering of Rhodococcus have also included the use of synthetic biology platforms and new approaches for genome editing (CRISPR/Cas9 and recombineering) mostly targeting R. opacus strain PD630. Despite that more efforts are needed to expand system and synthetic biology tools for genome-scale engineering of Rhodococcus species different from the model ones, the application of first metabolic engineering strategies and novel molecular toolkits has highlighted the great potential of engineered Rhodococcus spp. strains in biotechnological and industrial applications.
References
Alvarez HM, Herrero OM, Silva RA, Hernández MA, Lanfranconi MP, Villalba MS (2019) Insights into the metabolism of oleaginous Rhodococcus spp. Appl Environ Microbiol 85(18):1–12. https://doi.org/10.1128/AEM.00498-19
Alvarez HM, Silva RA, Herrero M, Hernández MA, Villalba MS (2013) Metabolism of triacylglycerols in Rhodococcus species: insights from physiology and molecular genetics. J Mol Biochem 2:2119–2130. https://doi.org/10.1007/s00253-012-4360-1
Anthony WE, Carr RR, Delorenzo DM et al (2019) Development of Rhodococcus opacus as a chassis for lignin valorization and bioproduction of high-value compounds. Biotechnol Biofuels 12(1):1–14. https://doi.org/10.1186/s13068-019-1535-3
Auffret M, Labbé D, Thouand G, Greer CW, Fayolle-Guichard F (2009) Degradation of a mixture of hydrocarbons, gasoline, and diesel oil additives by Rhodococcus aetherivorans and Rhodococcus wratislaviensis. Appl Environ Microbiol 75(24):7774–7782. https://doi.org/10.1128/AEM.01117-09
Bell KS, Philp JC, Aw DWJ, Christofi N (1998) A review: the genus Rhodococcus. J Appl Microbiol 85(2):195–210. https://doi.org/10.1046/j.1365-2672.1998.00525.x
Bugg TDH (2001) Oxygenases: mechanisms and structural motifs for O2 activation. Curr Opin Chem Biol 5(5):550–555. https://doi.org/10.1016/S1367-5931(00)00236-2
Busch H, Hagedoorn PL, Hanefeld U (2019) Rhodococcus as a versatile biocatalyst in organic synthesis. Int J Mol Sci 20(19):1–36. https://doi.org/10.3390/ijms20194787
Cai C, Xu Z, Xu M, Cai M, Jin M (2020) Development of a Rhodococcus opacus cell factory for valorizing lignin to muconate. ACS Sustain Chem Eng 8(4):2016–2031. https://doi.org/10.1021/acssuschemeng.9b06571
Cappelletti M, Fedi S, Honda K, Ohtake H, Turner R, Zannoni D (2010) Monooxygenases involved in the n-alkanes metabolism by Rhodococcus sp. BCP1: molecular characterization and expression of alkB gene. J Biotechnol 150:259. https://doi.org/10.1016/j.jbiotec.2010.09.150
Cappelletti M, Fedi S, Zampolli J et al (2016) Phenotype microarray analysis may unravel genetic determinants of the stress response by Rhodococcus aetherivorans BCP1 and Rhodococcus opacus R7. Res Microbiol 167(9–10):766–773. https://doi.org/10.1016/j.resmic.2016.06.008
Cappelletti M, Pinelli D, Fedi S, Zannoni D, Frascari D (2018) Aerobic co-metabolism of 1,1,2,2-tetrachloroethane by Rhodococcus aetherivorans TPA grown on propane: kinetic study and bioreactor configuration analysis . J Chem Technol Biotechnol 93(1):155–165. https://doi.org/10.1002/jctb.5335
Cappelletti M, Presentato A, Piacenza E, Firrincieli A, Turner RJ, Zannoni D (2020) Biotechnology of Rhodococcus for the production of valuable compounds. Appl Microbiol Biotechnol 104(20):8567–8594. https://doi.org/10.1007/s00253-020-10861-z
Cappelletti M, Presentato A, Milazzo G et al (2015) Growth of Rhodococcus sp. strain BCP1 on gaseous n-alkanes: new metabolic insights and transcriptional analysis of two soluble di-iron monooxygenase genes. Front Microbiol 6:1–15. https://doi.org/10.3389/fmicb.2015.00393
Cappelletti M, Zampolli J, Di Gennaro P, Zannoni D (2019) Genomics of Rhodococcus. In: Alvarez HM (ed) Biology of Rhodococcus second edition of the series Microbiology monographs. Springer, Heidelberg, p. 23–60. https://doi.org/10.1007/978-3-030-11461-9
Ceniceros A, Dijkhuizen L, Petrusma M, Medema MH (2017) Genome-based exploration of the specialized metabolic capacities of the genus Rhodococcus. BMC Genomics 18(1):1–16. https://doi.org/10.1186/s12864-017-3966-1
De Carvalho CCCR, Costa SS, Fernandes P, Couto I, Viveiros M (2014) Membrane transport systems and the biodegradation potential and pathogenicity of genus Rhodococcus. Front Physiol 5:1–13. https://doi.org/10.3389/fphys.2014.00133
DeLorenzo DM, Henson WR, Moon TS (2017) Development of chemical and metabolite sensors for Rhodococcus opacus PD630. ACS Synth Biol 6:1973–1978. https://doi.org/10.1021/acssynbio.7b00192
DeLorenzo DM, Moon TS (2019) Construction of genetic logic gates based on the T7 RNA polymerase expression system in Rhodococcus opacus PD630. ACS Synth Biol 8:1921–1930. https://doi.org/10.1021/acssynbio.9b00213
DeLorenzo DM, Rottinghaus AG, Henson WR, Moon TS (2018) Molecular toolkit for gene expression control and genome modification in Rhodococcus opacus PD630. ACS Synth Biol 7:727–738. https://doi.org/10.1021/acssynbio.7b00416
Desomer J, Dhaese P, Van Montagu M (1990) Transformation of Rhodococcus fascians by high-voltage electroporation and development of R. fascians cloning vectors. Appl Environ Microbiol 56(9):2818–2825. https://doi.org/10.1128/aem.56.9.2818-2825.1990
Dragosits M, Mattanovich D (2013) Adaptive laboratory evolution - principles and applications for biotechnology. Microb Cell Fact 12(1):1–17. https://doi.org/10.1186/1475-2859-12-64
Duran R (1998) New shuttle vectors for Rhodococcus sp. R312 (formerly Brevibacterium sp. R312), a nitrile hydratase producing strain. J Basic Microbiol 38(2):101–106
Gatti DL, Palfey BA, Lah MS, Entsch B, Massey V, Ballou DP, Ludwig ML (1994) The mobile flavin of 4-OH benzoate hydroxylase. Science 266(5182):110–4. https://doi.org/10.1126/science.7939628
van der Geize R, Dijkhuizen L (2004) Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr Opin Microbiol 7(3):255–261. https://doi.org/10.1016/j.mib.2004.04.001
Guevara G, Flores YO, De Las Heras LF, Perera J, Navarro Llorens JM (2019) Metabolic engineering of Rhodococcus ruber Chol-4: a cell factory for testosterone production. PLoS One 14(7). https://doi.org/10.1371/journal.pone.0220492
Henson WR, Campbell T, DeLorenzo DM, Gao Y, Berla B, Kim SJ, Foston M, Moon TS, Dantas G (2018) Multi-omic elucidation of aromatic catabolism in adaptively evolved Rhodococcus opacus. Metab Eng 49:69–83. https://doi.org/10.1016/j.ymben.2018.06.009
Hernández MA, Alvarez HM (2019) Increasing lipid production using an NADP+-dependent malic enzyme from Rhodococcus jostii. Microbiol (united Kingdom) 165(1):4–14. https://doi.org/10.1099/mic.0.000736
Hernández MA, Arabolaza A, Rodríguez E, Gramajo H, Alvarez HM (2013) The atf2 gene is involved in triacylglycerol biosynthesis and accumulation in the oleaginous Rhodococcus opacus PD630. Appl Microbiol Biotechnol 97(5):2119–2130. https://doi.org/10.1007/s00253-012-4360-1
Hernández MA, Comba S, Arabolaza A, Gramajo H, Alvarez HM (2015) Overexpression of a phosphatidic acid phosphatase type 2 leads to an increase in triacylglycerol production in oleaginous Rhodococcus strains. Appl Microbiol Biotechnol 99(5):2191–2207. https://doi.org/10.1007/s00253-014-6002-2
Hernández MA, Gleixner G, Sachse D, Alvarez HM (2017) Carbon allocation in Rhodococcus jostii RHA1 in response to disruption and overexpression of nlpR regulatory gene, based on 13C-labeling analysis. Front Microbiol 8:1–11. https://doi.org/10.3389/fmicb.2017.01992
Hernández MA, Mohn WW, Martínez E, Rost E, Alvarez AF, Alvarez HM (2008) Biosynthesis of storage compounds by Rhodococcus jostii RHA1 and global identification of genes involved in their metabolism. BMC Genomics 9:1–13. https://doi.org/10.1186/1471-2164-9-600
Hetzler S, Steinbüchel A (2013) Establishment of cellobiose utilization for lipid production in Rhodococcus opacus PD630. Appl Environ Microbiol 79:3122–3125. https://doi.org/10.1128/AEM.03678-12
Huang L, Zhao L, Zan X, Song Y, Ratledge C (2016) Boosting fatty acid synthesis in Rhodococcus opacus PD630 by overexpression of autologous thioesterases. Biotechnol Lett 38(6):999–1008. https://doi.org/10.1007/s10529-016-2072-9
Hwangbo M, Chu KH (2020) Recent advances in production and extraction of bacterial lipids for biofuel production. Sci Total Environ 734. https://doi.org/10.1016/j.scitotenv.2020.139420
Ivshina IB, Tyumina EA, Kuzmina MV, Vikhareva EV (2019) Features of diclofenac biodegradation by Rhodococcus ruber IEGM 346. Sci Rep 9(1):1–13. https://doi.org/10.1038/s41598-019-45732-9
Jiao S, Yu H, Shen Z (2018) Core element characterization of Rhodococcus promoters and development of a promoter-RBS mini-pool with different activity levels for efficient gene expression. N Biotechnol 44:41–49. https://doi.org/10.1016/j.nbt.2018.04.005
Kalscheuer R, Arenskötter M, Steinbüchel A (1999) Establishment of a gene transfer system for Rhodococcus opacus PD630 based on electroporation and its application for recombinant biosynthesis of poly(3-hydroxyalkanoic acids). Appl Microbiol Biotechnol 52(4):508–515. https://doi.org/10.1007/s002530051553
Kim HM, Chae TU, Choi SY, Kim WJ, Lee SY (2019) Engineering of an oleaginous bacterium for the production of fatty acids and fuels. Nat Chem Biol 15(7):721–729. https://doi.org/10.1038/s41589-019-0295-5
Kis Á, Laczi K, Zsíros S et al (2017) Characterization of the Rhodococcus sp. MK1 strain and its pilot application for bioremediation of diesel oil-contaminated soil. Acta Microbiol Immunol Hung 64(4):463–482. https://doi.org/10.1556/030.64.2017.037
Kis Á, Laczi K, Zsíros S, Rákhely G, Perei K (2015) Biodegradation of animal fats and vegetable oils by Rhodococcus erythropolis PR4. Int Biodeterior Biodegrad 105:114–119. https://doi.org/10.1016/j.ibiod.2015.08.015
Kurosawa K, Laser J, Sinskey AJ (2015a) Tolerance and adaptive evolution of triacylglycerol-producing Rhodococcus opacus to lignocellulose-derived inhibitors. Biotechnol Biofuels 8:76. https://doi.org/10.1186/s13068-015-0258-3
Kurosawa K, Plassmeier J, Kalinowski J, Rückert C, Sinskey AJ (2015b) Engineering L-arabinose metabolism in triacylglycerol-producing Rhodococcus opacus for lignocellulosic fuel production. Metab Eng 30:89–95. https://doi.org/10.1016/j.ymben.2015.04.006
Kurosawa K, Radek A, Plassmeier JK, Sinskey AJ (2015c) Improved glycerol utilization by a triacylglycerol-producing Rhodococcus opacus strain for renewable fuels. Biotechnol Biofuels 8:31. https://doi.org/10.1186/s13068-015-0209-z
Kurosawa K, Wewetzer SJ, Sinskey AJ (2013) Engineering xylose metabolism in triacylglycerol-producing Rhodococcus opacus for lignocellulosic fuel production. Biotechnol Biofuels 6:134. https://doi.org/10.1186/1754-6834-6-134
Kurosawa K, Wewetzer SJ, Sinskey AJ (2014) Triacylglycerol production from corn stover using a xylose-fermenting Rhodococcus opacus strain for lignocellulosic biofuels. J Microbial Biochem Technol 6:254–259. https://doi.org/10.4172/1948-5948.1000153
Laczi K, Kis Á, Horváth B et al (2015) Metabolic responses of Rhodococcus erythropolis PR4 grown on diesel oil and various hydrocarbons. Appl Microbiol Biotechnol 99(22):9745–9759. https://doi.org/10.1007/s00253-015-6936-z
Larcher S, Yargeau V (2011) Biodegradation of sulfamethoxazole by individual and mixed bacteria. Appl Microbiol Biotechnol 91(1):211–218. https://doi.org/10.1007/s00253-011-3257-8
Larkin MJ, Kulakov LA, Allen CCR (2005) Biodegradation and Rhodococcus - masters of catabolic versatility. Curr Opin Biotechnol 16(3 SPEC. ISS.):282–290. https://doi.org/10.1016/j.copbio.2005.04.007
Liang Y, Jiao S, Wang M, Yu H, Shen Z (2020) A CRISPR/Cas9-based genome editing system for Rhodococcus ruber TH. Metab Eng 57:13–22. https://doi.org/10.1016/j.ymben.2019.10.003
MacEachran DP, Sinskey AJ (2013) The Rhodococcus opacus TadD protein mediates triacylglycerol metabolism by regulating intracellular NAD(P)H pools. Microb Cell Fact 12(1):1. https://doi.org/10.1186/1475-2859-12-104
Mandal B, Prabhu A, Pakshirajan K, Veeranki Dasu V (2019) Construction and parameters modulation of a novel variant Rhodococcus opacus BM985 to achieve enhanced triacylglycerol-a biodiesel precursor, using synthetic dairy wastewater. Process Biochem 84(June):9–21. https://doi.org/10.1016/j.procbio.2019.05.031
Martínková L, Uhnáková B, Pátek M, Nešvera J, Křen V (2008) Biodegradation potential of the genus Rhodococcus. Environ Int 35(1):162–177. https://doi.org/10.1016/j.envint.2008.07.018
Mitani Y, Nakashima N, Sallam KI, Toriyabe T, Kondo K, Tamura T (2006) Advances in the development of genetic tools for the genus Rhodococcus. Actinomycetologica 20(2):55–61. https://doi.org/10.3209/saj.20.55
O’Brien EJ, Monk JM, Palsson BO (2015) Using genome-scale models to predict biological capabilities. Cell 161:971–987. https://doi.org/10.1016/j.cell.2015.05.019
Orro A, Cappelletti M, D’Ursi P et al (2015) Genome and phenotype microarray analyses of Rhodococcus sp. BCP1 and Rhodococcus opacus R7: genetic determinants and metabolic abilities with environmental relevance. PLoS One 10(10):1–41. https://doi.org/10.1371/journal.pone.0139467
Pátek M, Grulich M, Nešvera J (2021) Stress response in Rhodococcus strains. Biotechnol Adv. https://doi.org/10.1016/j.biotechadv.2021.107698
Presentato A, Cappelletti M, Sansone A et al (2018a) Aerobic growth of Rhodococcus aetherivorans BCP1 using selected naphthenic acids as the sole carbon and energy sources. Front Microbiol 9(APR):1–15. https://doi.org/10.3389/fmicb.2018.00672
Presentato A, Piacenza E, Anikovskiy M, Cappelletti M, Zannoni D, Turner RJ (2016) Rhodococcus aetherivorans BCP1 as cell factory for the production of intracellular tellurium nanorods under aerobic conditions. Microb Cell Factories 15:204. https://doi.org/10.1186/s12934-016-0602-8
Presentato A, Piacenza E, Anikovskiy M, Cappelletti M, Zannoni D, Turner RJ (2018b) Biosynthesis of selenium-nanoparticles and -nanorods as a product of selenite bioconversion by the aerobic bacterium Rhodococcus aetherivorans BCP1. New Biotechnol 41:1–8. https://doi.org/10.1016/j.nbt.2017.11.002
Presentato A, Piacenza E, Darbandi A, Anikovskiy M, Cappelletti M, Zannoni D, Turner RJ (2018c) Assembly, growth and conductive properties of tellurium nanorods produced by Rhodococcus aetherivorans BCP1. Sci Rep 8:3923. https://doi.org/10.1038/s41598-018-22320-x
Presentato A, Piacenza E, Turner RJ, Zannoni D, Cappelletti M (2020) Processing of metals and metalloids by actinobacteria: cell resistance mechanisms and synthesis of metal(loid)-based nanostructures. Microorganisms 8(12):1–37. https://doi.org/10.3390/microorganisms8122027
Roell GW, Carr RR, Campbell T et al (2019) A concerted systems biology analysis of phenol metabolism in Rhodococcus opacus PD630. Metab Eng 55(June):120–130. https://doi.org/10.1016/j.ymben.2019.06.013
Sandberg TE, Salazar MJ, Weng LL, Palsson BO, Feist AM (2019) The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng 56(August):1–16. https://doi.org/10.1016/j.ymben.2019.08.004
Schweizer E, Hofmann J (2004) Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol Mol Biol Rev 68(3):501–517. https://doi.org/10.1128/MMBR.68.3.501-517.2004
Sekizaki T, Tanoue T, Osaki M, Shimoji Y, Tsubaki S, Takai S (1998) Improved electroporation of Rhodococcus equi. J Vet Med Sci 60(2):277–279. https://doi.org/10.1292/jvms.60.277
Shao Z, Dick WA, Behki RM (1995) An improved Eschehchia coli‐Rhodococcus shuttle vector and plasmid transformation in Rhodococcus spp. using electroporation. Lett Appl Microbiol 21(4):261–266. https://doi.org/10.1111/j.1472-765X.1995.tb01056.x
Singer ME, Finnerty WR (1988) Construction of an Escherichia coli-Rhodococcus shuttle vector and plasmid transformation in Rhodococcus spp. J Bacteriol 170(2):638–645. https://doi.org/10.1128/jb.170.2.638-645.1988
Spence EM, Calvo-Bado L, Mines P, Bugg TDH (2021) Metabolic engineering of Rhodococcus jostii RHA1 for production of pyridine-dicarboxylic acids from lignin. Microb Cell Fact 20(1):1–12. https://doi.org/10.1186/s12934-020-01504-z
Tajparast M, Frigon D (2015) Genome-scale metabolic model of Rhodococcus jostii RHA1 (iMT1174) to study the accumulation of storage compounds during nitrogen-limited condition. BMC Syst Biol 9:43. https://doi.org/10.1186/s12918-015-0190-y
Tajparast M, Frigon D (2018) Predicting the accumulation of storage compounds by Rhodococcus jostii RHA1 in the feast-famine growth cycles using genome-scale flux balance analysis. PLoS One 13
Tyumina EA, Bazhutin GA, Vikhareva EV, Selyaninov AA, Ivshina IB (2019) Diclofenac as a factor in the change of Rhodococcus metabolism. IOP Conf Ser Mater Sci Eng 487(1). https://doi.org/10.1088/1757-899X/487/1/012027
Weidhaas JL, Chang DPY, Schroeder ED (2009) Biodegradation of nitroaromatics and RDX by isolated Rhodococcus opacus. J Environ Eng 135(10):1025–1031. https://doi.org/10.1061/(asce)ee.1943-7870.0000072
Xie S, Sun S, Lin F et al (2019) Mechanism-guided design of highly efficient protein secretion and lipid conversion for biomanufacturing and biorefining. Adv Sci 6(13). https://doi.org/10.1002/advs.201801980
Xiong X, Lian J, Yu X, Garcia-Perez M, Chen S (2016a) Engineering levoglucosan metabolic pathway in Rhodococcus jostii RHA1 for lipid production. J Ind Microbiol Biotechnol 43:1551–1560. https://doi.org/10.1007/s10295-016-1832-9
Xiong X, Wang X, Chen S (2012) Engineering of a xylose metabolic pathway in Rhodococcus spp. strains. Appl Environ Microbiol 78:5483–5491. https://doi.org/10.1128/AEM.08022-11
Xiong X, Wang X, Chen S (2016b) Engineering of an L-arabinose metabolic pathway in Rhodococcus jostii RHA1 for biofuel production. J Ind Microbiol Biotechnol 43:1017–1025. https://doi.org/10.1007/s10295-016-1778-y
Yoneda A, Henson WR, Goldner NK, Park KJ, Forsberg KJ, Kim SJ, Pesesky MW, Foston M, Dantas G, Moon TS (2016) Comparative transcriptomics elucidates adaptive phenol tolerance and utilization in lipid-accumulating Rhodococcus opacus PD630. Nucleic Acids Res 44:2240–2254. https://doi.org/10.1093/nar/gkw055
Acknowledgements
The authors are greatly thankful to the anonymous reviewers for the useful comments and suggestions that helped in improving the final version of the manuscript.
Funding
Open access funding provided by Alma Mater Studiorum - Università di Bologna within the CRUI-CARE Agreement. RFO Grant University of Bologna (RFO Grant UNIBO).
Author information
Authors and Affiliations
Contributions
E.D. performed the literature search and wrote the first draft of the manuscript; A. F. contributed to the writing of the paragraphs “Genome-scale metabolic modeling” and “Adaptive Laboratory Evolution”; M.C. conceptualized the manuscript, supervised the writing, and critically revised and edited the entire manuscript.
Corresponding author
Ethics declarations
Consent to participate
All the authors consent to participate.
Consent for publication
All the authors consent to publish the manuscript.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Donini, E., Firrincieli, A. & Cappelletti, M. Systems biology and metabolic engineering of Rhodococcus for bioconversion and biosynthesis processes. Folia Microbiol 66, 701–713 (2021). https://doi.org/10.1007/s12223-021-00892-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-021-00892-y