Abstract
DNA methylation is an essential epigenetic modification involved in numerous biological processes. Here, we present a cell-based system pLTR-Luc2P-EGFP for evaluation of DNA methylation in mammalian cells. In this system, the expression of reporter gene luciferase2P (Luc2P)-EGFP is under the control of HIV-1 promoter 5' long terminal repeat (LTR), which contains multiple CpG sites. Once these sites are methylated, the expression of Luc2P-EGFP is turned off, which may be visualized under fluorescence microscopy, with quantification performed in luciferase activity assay. As a proof of principle, pLTR-Luc2P-EGFP was methylated in vitro, and transfected into 293T cells, where the reduction of Luc2P-EGFP expression was confirmed. Premixed reporter DNA samples with the methylation levels varying from 0 to 100% were used for quantitative measurements of DNA methylation. The resulting standard curves indicated the accuracy of luciferase activity exceeding that of the Western blotting against EGFP. The Bland–Altman analysis showed that data from luciferase activity assay were in good agreement with the actual DNA methylation levels. In summary, we have established a reporter system coupled with reliable detection technique capable of efficient quantifying the changes in methylation in mammalian cells. This system may be utilized as a high throughput screening tool for identifying molecules that modulate DNA methylation.
Similar content being viewed by others

INTRODUCTION
Epigenetics describes the phenotypic changes that alter gene expression without disturbing the primary DNA sequences [1]. Epigenetic regulations are heritable and reversible; their list includes DNA methylation, histone modifications and small noncoding microRNAs (miRNA) [1, 2]. Since Riggs [3] and Holiday et al. [4] proposed a convincing model of molecular mechanism of inheritance more than 40 years ago, DNA methylation had been considered as a paradigm of epigenetic information transfer [5]. Currently, DNA methylation is a common epigenetic modifications in eukaryotes, which plays an important regulatory role in biological processes such as transposable element silencing, genomic imprinting, X chromosome inactivation and developmental processes [6, 7], and abnormal methylation patterns are often associated with the incidence of diseases [8].
In mammals, most of the methylations occur at the carbon-5 position of cytosine (5 mC) [9–11]. The methylcytosine is mainly found in cytosine-guanine (CpG) dinucleotides. Although the CpG dinucleotides constitute only 1% of the human genome, CpG-rich stretches, so-called CpG islands, are located in the promoter regions of more than 70% of all known human genes [12–14]. 5mCs, especially when clustered at CpG sites, are important transcriptional silencers at gene promoters and endogenous retrotransposons in the genome [15–17]. Many studies have shown that, the epigenetic silencing of a variety of genes by hyper methylation of promoter-associated CpG islands is often related with particular diseases [18–20]. Therefore, a simple, reliable and sensitive method for detecting DNA methylation and its changes (e.g. hyper- or hypomethylation) is of great interest.
The traditional DNA methylation assays are mainly based on sodium bisulfite treatment, which converts non-methylated cytosine into uracil, while methylated cytosine is resistant to bisulfite and remains unchanged [21]. This allows discrimination between methylated DNA and non-methylated DNA, and usually followed by methylation-specific PCR, DNA sequencing or combined bisulfite restriction assays. Dues to their reliability and accuracy, they are widely used to quantify the site specific DNA methylation. However, these assays require complex procedures such as cloning and sequencing, which limits their usage in high-throughput analysis [22, 23]. Meanwhile, techniques based on high-performance liquid chromatography(HPLC) [24], restriction enzyme PCR [25] and gas chromatography/mass spectrometry (GC/MS) [26] have been developed. These techniques are complex, time consuming and expensive. Therefore, the more convenient and easy to use assay assessing DNA methylation warrants development.
Luciferase is a type of bioluminescence producing enzymes isolated from several animal species. Due to the high sensitivity, robust signal and assay convenience, it is often used as reporter to monitor gene activity/gene promoter activity. For example, Sanchez et al. used the luciferase reporter gene to analyze the relationship between the structure and biological activity of strigolactones [27]. Solberg et al used a luciferase assay to characterize the activity of the 5'‑flanking promoter of mouse Tcf3 [28]. Moreover, the combination of two luciferases: firefly luciferase and Renilla luciferase, improved the quantitative accuracy and assay reproducibility, where Renilla luciferase serves as an internal control to monitor cell numbers and viability. So the luciferase reporter assay is a powerful tool used to study the regulatory elements of genes of interest. For example, Xiao et al. established the 293-Sox2-Luciferase cell line as a luciferase reporter system to study transcriptional regulation of the human Sox2 gene [29]. And recently, a luciferase reporter virus icSARS-CoV-2-nLuc-GFP was established to test the cross-CoV neutralization of sera from SARS and COVID-19 patients [30].
The epigenetic regulation theory suggest that the promoter activity is consistent with the level of DNA methylation [31, 32]. Therefore, the luciferase was further used as a reporter to reflect the methylation status and/or methylation changes of gene promoter. For example, Li et al. used a cell-based firefly luciferase reporter assay to test the effect of Gadd45a on DNA methylation [33]. And the evaluation of CpG methylation by using HIV LTR-luciferase plasmid was mentioned in a short report [34]. However, the quantitative accuracy and reliability of the detection, including the dose-response relationship between methylation levels and luciferase activity, remain to be assessed.
In order to fully characterize the properties of luciferase-based methylation reporter system, here we used a modified firefly luciferase gene Luc2P fused with EGFP as the reporter to represent the promoter activity, and established a cell based system to measure the changes of DNA methylation. The result from luciferase activity assay was validated by comparing it with the data obtained by HpaII sensitivity assay [35] and Western blotting analysis [36, 37]. After carefully analysis, we proved that this is a reliable reporter system for accurate measuring the changes of DNA methylation in living cells.
EXPERIMENTAL
DNA constructions. The EGFP expression plasmid pEGFP-N1 was purchased from BD Biosciences Clontech, and used as backbone for pLTR-Luc2P-EGFP construction. The HIV-1 5' LTR was PCR amplified from HIV-1 pNL4.3, and used to replace the CMV promoter of pEGFP-N1 by AseI and NheI digestion. Luc2P was amplified from pGL4.32 [luc2P/NFkB-RE/Hygro] (Promega), and cloned into pEGFP-N1 within HindIII and BamHI sites. The constructed plasmid pLTR-Luc2P-EGFP was verified by sequencing. Primers used for PCR amplification are: 5'-AseI-LTR, TCGTATTAATTGGAAGGGCTAATTTGGTC, 3'-NheI-LTR, CTAGCTAGCTGCTAGAGATTTTCCAC-ACTGAC, 5'-HindIII-luc2P CCCAAGCTTATGGAAGATGCCAAAAACATTA, 3'-BamHI-luc2P, CGGGATCCGACGTTGATCCTGGCGCTGG.
In vitro methylation of plasmid DNA. The plasmid pLTR-Luc2P-EGFP was treated with CpG methylase M.SssI (Zymo). Briefly, 2 μg plasmid was incubated with 0.6 mM S-adenosylmethionine and 4 units M.SssI at 30°C for 6 h, then 2 units of M.SssI were added to the reaction system and continuing incubated at 30°C for 6–8 h. The treated plasmid was recycled by 3 M sodium acetate. Complete CpG methylation of the pLTR-Luc2P-EGFP was confirmed by endonucleases (HpaII and MspI, Thermo) digestion and bisulfite-mediated methylcytosine mapping.
Bisulfite-mediated methylcytosine mapping. After bisulfite conversion (Zymo), regions of interest in M.SssI treated plasmid pmeLTR-Luc2P-EGFP and non-methylated plasmid pLTR-Luc2P-EGFP were amplified by PCR (primers targeting LTR: LTR-BSP-F, 5'-TATGAGTTAGTATGGGATGGAGGAT-3', and LTR-BSP-R, 5'-AATCTAACCAAAAAAACCCAA-TACA-3'). The PCR products were gel-purified by using DNA Gel Extraction Kit (Axygen) and cloned into pJET1.2 for sequencing. Data were analyzed by online software QUantification tool for Methylation Analysis (http://quma.cdb.riken.jp/) [38].
Cell culture and transient transfection. Human embryonic kidney (HEK) 293T cells and HeLa cells were maintained in DMEM (Gibco) supplemented with 100 units/mL penicillin, 0.1 mg/mL streptomycin, and 10% fetal bovine serum (FBS, Gibco). Cells were cultured at 37°C in 5% CO2 incubator.
For transient transfection, 293T cells were plated onto 12-well plates. When about 80% confluent, cells were transfected with ummethylated pLTR-Luc2P-EGFP or methylated pmeLTR-Luc2P-EGFP plasmids using FuGENE® HD reagent (Promega), following manufacturer’s protocol. The Renilla luciferase control vector pGL4.74 [hRluc/TK] (Promega) was co-transfected with reporter vector at the ratio of 1 : 10.
Immunofluorescence assay. For the detection of 5mC, HeLa cells grown on coverslips were transfected with the ummethylated and methylated pLTR-Luc2P-EGFP (0.2 μg/well) for 36 hours and fixed in 4% paraformaldehyde in PBS for 15 min at room temperature. The cells were rinsed with PBS thrice and incubated in PBS containing 5% fetal bovine serum and 0.3% Triton X-100 for blocking, and then incubated overnight at 4°C with 5-methylcytosine monoclonal antibodies (1 : 100, EpiGentek). The next day after rewarming at RT for 1 h, the cells were washed thrice with PBS, and incubated with goat anti-mouse IgG (H + L) coupled with Alexa Fluor 555 dyes antibody (1 : 500, Thermo) for 1 h at 37°C. After three washes with PBS, the cells were incubated with DAPI staining solution (Beyotime) for 3–5 min. Following three washes with PBS, the samples were mounted on slides with anti-fade Fluorescence Mounting Medium (Beyotime) and examined with a confocal microscope (Leica Inc, Germany).
For the detection of EGFP, 36–48 hrs post-transfection, 293 T cells with EGFP fluorescence signal were observed under Nikon fluorescence inverted microscope (Nikon ECLIPSE Ti), images were captured and processed with NIS-Elements D.
Western blotting. 48 hrs post transfection, 293T cells were lysed on ice in RIPA buffer plus protease inhibitors (Beyotime, China). Lysates were fractionated by SDS-PAGE and transferred to nitrocellulose (NC) membrane (Millipore). Membranes were blocked with 5% nonfat dry milk in PBS, incubated with rabbit anti-GFP antibody (1/400 dilution in PBS, Santa Cruz), and again incubated with secondary antibody goat anti-rabbit IgG-HRP (1/10000 dilution in PBS, Elabscience), GAPDH immunoblotting was performed with anti-human-GAPDH antibody (1 : 10000, Sigma) as control. Detection was performed using enhanced chemiluminescence (Bio-Rad).
Luciferase activity assay. The Dual-Glo® Luciferase Reporter Assay System (Promega) was used for the firefly and Renilla luciferase activity measurement. Briefly, 48-h post transfection, Dual-Glo® Reagent equal to the volume of culture medium was added to the plate wells. The plate was subjected to end-over-end rotation for 20 min to achieve complete lysis. And the firefly luminescence (Fluc) was measured in a GloMax®-96 microplate luminometer (Promega). Then, equal amount of Dual-Glo® Stop & Glo® Reagent was added, and the Renilla luminescence (Rluc) was measured 20 min later. The Relative Luciferase Activity (RLA) was calculated by dividing the Fluc by Rluc. Relative Response Ratio (RRR) was calculated by [(experimental RLA) – (RLA of cells transfected with 100% methylated DNA)]/[(RLA of cells transfected with 0% methylated DNA) – (RLA of cells transfected with 100% methylated DNA)].
HpaII sensitivity assay. The CpG methylation levels of 5'-LTR in pLTR-Luc2P-EGFP were quantified using the quantitative real-time PCR(qPCR) after HpaII digestion, which is blocked by CpG methylation [35]. Total DNA was extracted from transfected cells with Quick-DNATM Miniprep Plus Kit (Zymo). 0.5 μg DNA was incubated with 10 units HpaII or in a mock reaction without HpaII at 37°C for 4 h, and inactivated for 20 min at 80°C. Equal amounts from both the mock reaction and the HpaII reaction were used in qPCR using the SYBR Premix Ex Taq (Tli RNaseH Plus, Takara). 5'-LTR flanking two HpaII digestion sites was amplified. Housekeeping gene GAPDH was used as the internal reference. PCR reaction was run on an Applied Biosystems 7500 Real-Time PCR system under the following conditions: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Primer sequences for 5'-LTR are: forward-GCCAATGAAGGAGAGAACAACA; reverse-AGCGGAAAGTCCCTTGTA; for GAPDH are: forward-GAAGGTGAAGGTCGGAGTCAAC and reverse-CAGAGTTAAAAGCAGCCCTGGT. Reaction was run in triplicate. HpaII sensitivity of the CCGG site was calculated by [1–2Ct (mock) – Ct (HpaII)] × 100%.
Statistical analysis. Data are shown as the mean ± SD of at least two independent experiments; each replicate has at least three technical replicates. The two-tailed Student’s t-test and one-way analysis of variance (ANOVA) were performed using SPSS (version 21, IBM). A value of P < 0.05 was considered statistically significant.
Histogram was obtained by Graphpad Prism 6 to show the data in the most intuitive way. The standard curve was obtained by Excel 2013 to show correlation between three DNA methylation assays and standard. Bland–Altman plot was obtained by MedCalc software to analysis the consistency of three DNA methylation assays.
RESULTS
Construction and Methylation of the Reporter Vector pLTR-Luc2P-EGFP
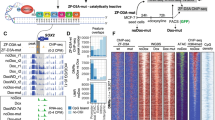
To construct the report vector, pEGFP-N1 was used as the backbone. The modified firefly luciferase Luc2P was inserted into multiple cloning site, and the fusion protein Luc2P-EGFP was used as the reporter. The original promoter CMV was removed and replaced with HIV-1 5'LTR, which contains eight CpG sites and is easily methylated [34, 39] (Fig. 1a). After methylated in vitro by M.sssI, the pmeLTR-Luc2P-EGFP can be delivered into mammalian cells. Then the methylation status of the promoter 5'LTR can be monitored by HpaII sensitivity assay (real time-PCR) directly [35], or indirectly reflected by the expression of the reporter gene Luc2P-EGFP. In transfected cells, the expression of Luc2P-EGFP can be qualitatively monitored by fluorescence microscopy, semi-quantitative monitored by Western blotting (anti-GFP antibody) or quantified by luciferase activity assay (Fig. 1b).
Overview of reporter vector and experimental procedures. (a) Schematic representation of the pLTR-Luc2P-GFP construction, key components were shown, in which the expression of Luc2P-EGFP fusion protein was under the control of HIV-1 5'LTR promoter. (b) A schematic illustration of the experimental procedure.
After M.sssI treatment, pmeLTR-Luc2P-EGFP was analyzed by HpaII/MspI digestion. Both enzymes recognize the same site-CCGG, but the HpaII digestion can be blocked by CpG methylation [40]. As shown in Fig. 2a, both methylated and non-methylated DNA could be cleaved by MspI, which confirmed the presence of CCGG sites in pLTR-Luc2P-EGFP. But only M.sssI treated DNA was resistant to the HpaII digestion, indicating the CpG sites in pmeLTR-Luc2P-EGFP were fully methylated by M.sssI treatment. Also, the methylation efficiency was confirmed by the bisulfite sequencing of LTR region. As shown in Fig. 2b, among the six M.sssI treated plasmid clones (M1–M6), the methylation rate of CpG sites was 87.5% in four clones, and 75% in the other two clones. In contrast, 0% of CpG sites were methylated in all six non-methylated clones (U1–U6). Then pmeLTR-Luc2P-EGFP was transfected into HeLa cells, the presence of 5mC were measured by immunofluorescence assay. In the cells transfected with non-methylated DNA, the presence of 5mC was not detected (Fig. 2c, lower panel). In pmeLTR-Luc2P-EGFP transfected cells, 5mC was detected and mainly located in the nucleus (Fig. 2c, upper panel), suggesting that, pmeLTR-Luc2P-EGFP remains methylated and stable after delivered into living cells.
Analysis of the M.SssI mediated DNA methylation. (a) DNA gel image of restriction endonuclease digestion. Methylated (M.SssI+) and non-methylated (M.SssI–) pLTR-Luc2P-GFP DNA substrates were digested as shown. HM, CCC, and Cut indicate higher mass, covalently closed circular and cleaved plasmid respectively. (b) Bisulfite-mediated mapping of pLTR-Luc2P-GFP. Each row represents an independently cloned DNA molecule from methylated (M1–M6) or non-methylated (U1–U6) pLTR-Luc2P-EGFP. Each column represents one CpG within LTR region. Open-circles indicate non-methylated CpG sites, while closed-circles indicate methylated CpG sites. The methylation level is presented as percentage of methylated CpGs in each clone. (c) Immunofluorescent analysis of 5mC in methylated (M) or non-methylated (UM) pLTR-Luc2P-EGFP transduced cells. HeLa cells transduced with reporter plasmid were costained for 5mC antibody and DAPI. The results are representative of the three experiments.
Validation of the Reporter System
In order to evaluate the reporter system, equal amounts of M.SssI treated or untreated reporter plasmids were delivered into 293T cells. 48-h following transfection, the cells are assayed by methods listed in Fig. 1b. Compared with cells transfected with non-methylated plasmid, strong silencing of Luc2P-GFP expression was observed in pmeLTR-Luc2P-EGFP transfected cells, as examined by fluorescence microscopy and Western blotting (Figs. 3a, 3b). Meanwhile, similar result was obtained by luciferase activity assay, that is, the relative luciferase activity of the pmeLTR-Luc2P-EGFP transfected cells was significantly reduced (Fig. 3c, P < 0.01). Moreover, the HpaII sensitivity of DNA isolated from pmeLTR-Luc2P-EGFP transfected cells was significantly decreased (Fig. 3d, P < 0.01), confirmed that the gene silencing is in consistence with the promoter methylation. These data showed that these three methods (by detecting EGFP, Luc2P, 5mC in LTR region) are capable of efficient distinguishing methylated DNA samples from non-methylated ones.
Validation of the reporter system pLTR-Luc2P-EGFP in 293T cells. (a) The fluorescence of Luc2P-EGFP under fluorescence microscopy. The fluorescence intensity of EGFP in 293T cells transfected with M or UM reporter vectors (left). Cells were plated at approximately equal density, as evidenced by phase contrast microscopy (right). (b) Western blotting analysis of Luc2P-EGFP in M and UM transduced cells. The blot plot is a representative of three experiments. The data of the histogram was determined by relative intensity using image analysis, and presented as mean ± SD from triplicate experiments. **P < 0.01, Student’s t-test. (c) The relative luciferase activity of 293T cells transfected with M and UM plasmid. The experiments were done in triplicate, with cultures repeated three times in total. Data are presented as mean ± SD. **P < 0.01, Student’s t-test. (d) The HpaII sensitivity of the LTR region in M and UM plasmid transfected cells. Open circle represents data from individual experiment and short line represents mean values. **P < 0.01, Student’s t-test.
Quantification of Methylation Status by the Reporter System
To determine whether this system allows quantitation of DNA methylation, we performed a dilution experiment. Fully methylated plasmid (pmeLTR-Luc2P-EGFP) was set as 100% and non-methylated plasmid (pLTR-Luc2P-EGFP) as 0%. The plasmids were mixed by different ratios (3 : 1, 2 : 2, 1 : 3) to yield 75, 50 and 25% methylated plasmid. DNA mix with different methylation gradients (100, 75, 50, 25, 0%) were transfected 293T cells, the methylation status were detected by corresponding methods. As shown in Fig. 4, decreasing of methylation levels result in a gradually increase of Luc2P-GFP expression, as indicated by fluorescence intensity, Western blotting, relative response ratio (RRR) of luciferase assay. Consistently with protein expression, HpaII sensitivity assay showed the same trend. Hereby, we confirmed that presented reporter system may find its use in quantitative measurements of DNA methylation.
Quantitative assessment of the reporter system accuracy. 293T cells transfected with pLTR-Luc2P-EGFP methylated at different levels (100, 75, 50, 25, 0%) were observed under fluorescence microscopy (a) and analyzed by Western blotting (b), luciferase activity assay as indicated by relative response radio (c) and HpaII sensitivity assay (d). The experiments were done in triplicate, with cultures repeated three times in total. Data of the histogram are presented as mean ± SD. The fluorescence image and blot plot is a representative of the three experiments. Open circles in (d) represent data from individual experiments and lines represent mean values. *P < 0.05, **P < 0.01, one-way analysis of variance (ANOVA).
Accuracy and Reliability Assessment
To evaluate quantitative accuracy of the detection methods, data from Fig. 4, plotted against their respective methylation percentage, were subjected to linear regression analysis. Figure 5 revealed a strong linear regression between normalized data points and the percentage of methylated DNA (R2 = 0.9156 for Western blotting; R2 = 0.9613 for luciferase assay; R2 = 0.9867 for HpaII sensitivity assay), suggesting a good correlation between the measured value and actual methylation level.
Linear regression analysis between the methylated level of transfected plasmid and normalized data points from Western blotting (a), luciferase activity assay (b) and HpaII sensitivity assay (c) of Fig. 4. Solid circle represents mean values, straight line represents the corresponding standard curve, broken line represents the connection between actual values. The equation of the linear regression curve and the correlation coefficient (R 2) are indicated.
The consistency of the measurement was further analyzed by Bland–Altman plot [41, 42]. All data points collected by Western blotting, luciferase assay and HpaII sensitivity assay are within the consistency limit (Fig. 6), namely 100% differences are within the consistency limit. It is generally believed that, if more than 95% of the differences are within the consistency limit, the consistency is considered to be good [42]. Therefore, the consistency of three assays was all in good agreement with the actual 5mC level of pLTR-Luc2P-EGFP.
Bland–Altman plots of Western blotting (a), luciferase activity assay (b) and HpaII sensitivity assay (c). D values represent the differences between values measured by the assay and their respective methylation percentage. The dotted lines are mean (%), the solid lines are mean (%) ± 1.96 SD, and the hollow circles represent the differences.
DISCUSSION
Here we present a reporter system for assessing the 5mC modification in mammalian cells coupled with accurate and reliable assay for visualization and quantitation of these methylation events including real time-PCR, Western blotting and luciferase activity assay. Despite that the first two methods only require resources commonly available in a laboratory of molecular biology, they are more cumbersome and time-consuming than luciferase activity assay. Luciferase activity assay requires specialized equipment and reagents, but does not require the sample preparation step. Therefore, it is rapid in operation, plus very sensitive in detection and easy to analysis, which enables researchers to obtain accurate results with minimal effort.
Luciferase was first found from firefly (Photinus pyralis) by de Wet J.R. et al. in 1985, and advanced in detection technology across academia and industry [43]. Currently, luciferase has been widely used in reporter system for functional genomics (such as RNAi screening), signaling pathways, well-defined molecular mechanism, and biological activity studies. While searching for the term “luciferase reporter assay”, over 20 000 publications since 1987 appeared [44]. As a reporter, luciferase genes have the following important features: exceptional sensitivity (10- to 1000-fold higher than fluorescent reporters such as GFP), wide dynamic range, typically no endogenous activity in host cells to interfere with quantitation, and, the measurements are almost instantaneous [45].
Therefore, here we used a dual-luciferase assay to evaluate the methylation of HIV-1 promoter 5' LTR in transfected 293T cells. Firefly luciferase Luc2P tracks the transcriptional activity of LTR promoter, while the Renilla luciferase acts as an internal control to minimize experimental variability due to pipetting errors, cell viability and transfection efficiency [46]. The ratio of two luciferase signals (relative luciferase activity) represents the relative expression the Luc2P-EGFP, and indirectly reflects the methylation status of the promoter. In our study, the expression of Luc2P-EGFP was also detected by Western blotting. The results revealed that, both methods could monitor protein expression with the linear range from 0 to 100% methylated DNA. While compared with Western blotting (R2 = 0.9156), luciferase assay has a higher accuracy (R2 = 0.9613). Meanwhile, we used the HpaII sensitivity assay to directly examine the methylation of promoter LTR, which is based on the combination of methylation sensitive restriction enzyme HpaII and qPCR [35, 47]. The results showed that, the HpaII sensitivity was also negatively correlated with the methylation level of pLTR-Luc2P-EGFP. And the Bland-Altman analysis revealed that, the consistency of luciferase activity assay was similar to HpaII sensitivity assay, and, in good agreement with the actual DNA methylation level. Thus, these data proved that, the luciferase activity assay is a valid substitute for detection of DNA methylation in living cells.
Moreover, the simplicity and sensitivity of luciferase activity assay makes it a high throughput screening tool for identification of novel compounds that modulate DNA methylation. In fact, luciferase assays have been used for screening of antimicrobial agents against Mycobacterium tuberculosis and immunosuppressive drugs as earlier as 1990s [48, 49]. After that, increasing studies proved the potential usefulness of luciferase assay for the screening. Since this method could not only reduce the cost of drug screening, but also improve the reliability and predictability of the results [50].
In terms of our reporter system, where luciferase is driven by HIV-1 promoter, it can be used to screen compounds targeting HIV-1 latency reactivation. HIV-1 latency is the major barrier for HIV-1 eradication in infected individuals. And reactivation of latent virus is the first step in the “kick and kill” strategy, a novel direction in HIV-1 cure [51]. DNA methylation in LTR region has been shown to be highly associated with latency regulation [52]. Hypermethylation suppresses viral gene expression and stabilizes HIV-1 latency. Compounds demethylating LTR reactivate latent virus, and can be further used in eliminating or reducing viral reservoirs [51, 53]. Therefore, instead of using HIV-infected cell lines or primary cells, where latency may be achieved by more than one mechanism, compounds reactivating virus solely via DNA demethylation may be screened in our reporter system, which is also applicable to the studies of LTR methylation in HIV-1 latency. Hence, the screens for therapeutic compounds against diseases caused by abnormal methylation changes may be broadened [54].
In conclusion, a luciferase-based reporter system pLTR-Luc2P-EGFP was developed and validated. Feasibility and reliably of this system for assessment of methylation changes is proven. This study promotes the use of bioluminescence in the field of epigenetics, and provides new method for screening compounds targeting DNA methylation/demethylation.
REFERENCES
Kanwal R., Gupta S. 2012. Epigenetic modifications in cancer. Clin. Genet. 81 (4), 303– 311.
Ellis L., Atadja P.W., Johnstone R.W. 2009. Epigenetics in cancer: targeting chromatin modifications. Mol. Cancer Ther. 8 (6), 1409–1420.
Riggs A.D. 1975. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 14, 9–25.
Holliday R., Pugh J.E. 1975. DNA modification mechanisms and gene activity during development. Science. 187, 226–232.
Jeltsch A., Jurkowska R.Z. 2014. New concepts in DNA methylation. Trends Biochem. Sci. 39 (7), 310–318.
Wu S.C., Zhang Y. 2010. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11 (9), 607–620.
Jaenisch R., Bird A. 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 33 (Suppl.), 245–254.
Nadasi E., Clark J.S., Szanyi I., Varjas T., Ember I., Baliga R., Arany I. 2009. Epigenetic modifiers exacerbate oxidative stress in renal proximal tubule cells. Anticancer Res. 29 (6), 2295–2299.
Rodriguez-Paredes M., Esteller M. 2011. Cancer epigenetics reaches mainstream oncology. Nat. Med. 17 (3), 330–339.
Zhu J.K. 2009. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43, 143–166.
Suzuki M.M., Bird A. 2008. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 9 (6), 465–476.
Momparler R.L. 2003. Cancer epigenetics. Oncogene. 22 (42), 6479–6483.
Plass C. 2002. Cancer epigenomics. Hum. Mol. Genet. 11 (20), 2479–2488.
Tsai A.G., Lu H., Raghavan S.C., Muschen M., Hsieh C.L., Lieber M.R. 2008. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 135 (6), 1130–1142.
Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E., Absher D.M., Wold B.J., Myers R.M. 2013. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23 (3), 555–567.
Baylin S.B., Jones P.A. 2011. A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer. 11 (10), 726–734.
Berman B.P., Weisenberger D.J., Aman J.F., Hinoue T., Ramjan Z., Liu Y., Noushmehr H., Lange C.P., van Dijk C.M., Tollenaar R.A., van Den Berg D., Laird P.W. 2011. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 44 (1), 40–46.
Xiwei W., Rauch T.A., Xueyan Z., Bennett W.P., Latif F., Krex D., Pfeifer G.P. 2010. CpG island hypermethylation in human astrocytomas. Cancer Res. 70 (7), 2718–2727.
Tao L.H., Wang W., Li L., Kramer P.K., Pereira M.A. 2005. DNA hypomethylation induced by drinking water disinfection by-products in mouse and rat kidney. Toxicol. Sci. 87 (2), 344–352.
Boultwood J., Wainscoat J.S. 2007. Gene silencing by DNA methylation in haematological malignancies. Br. J. Haematol. 138 (1), 3–11.
Fuso A., Scarpa S., Grandoni F., Strom R., Lucarelli M. 2006. A reassessment of semiquantitative analytical procedures for DNA methylation: Comparison of bisulfite- and HpaII polymerase-chain-reaction-based methods. Anat. Biochem. 350 (1), 24–31.
Aichinger E., Köhler C. 2010. Bisulphite sequencing of plant genomic DNA. Methods Mol. Biol. 655, 433–443.
Docherty S.J., Davis O.S.P., Haworth C.M.A., Plomin R., Mill J. 2010. DNA methylation profiling using bisulfite-based epityping of pooled genomic DNA. Methods. 52 (3), 255–258.
Kuo K.C., McCune R.A., Gehrke C.W., Midgett R., Ehrlich M. 1980. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 8 (20), 4763–4776.
Boers R., Boers J., de Hoon B., Kockx C., Ozgur Z., Molijn A., van IjckenW., Laven J., Gribnau J. 2018. Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 28 (1), 88–99.
Tang Y., Gao X.-D., Wang Y.S., Yuan B.-F., Feng Y.-Q. 2012. Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry. Analyt. Chem. 84 (16), 7249–7255.
Sanchez E., Artuso E., Lombardi C., Visentin I., Lace B., Saeed W., Lolli M.L, Kobauri P., Ali Z., Spyrakis F., Cubas P., Cardinale F., Prandi C. 2018. Structure-activity relationships of strigolactones via a novel, quantitative in planta bioassay. J. Exp. Bot. 69 (9), 2333–2343.
Solberg N., Machon O., Krauss S. 2012. Characterization and functional analysis of the 5'-flanking promoter region of the mouse Tcf3 gene. Mol. Cell. Biochem. 360, 289–299.
Xiao D., Zhang W., Li Y., Liu K., Zhao J., Sun X., Shan L., Mao Q., Xia H. 2016. A novel luciferase knock-in reporter system for studying transcriptional regulation of the human Sox2 gene. J. Biotechnol. 219, 110–116.
Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist Sarah R., et al. 2020. SARS-CoV-2 reversegenetics reveals a variable infection gradient in the respiratory tract. Cell. 182 (2), 429–446, e414.
Antequera F. 2003. Structure, function and evolution of CpG island promoters. Cell. Mol. Life Sci. 60 (8), 1647–1658.
Jones P.A. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13 (7), 484–492.
Li Z., Gu T.P., Weber A.R., Shen J.Z., Li B.Z., Xie Z.G., Yin R., Guo F., Liu X., Tang F., Wang H., Schar P., Xu G.-L. 2015. Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Res. 43 (8), 3986–3997.
Ishida T., Hamano A., Koiwa T., Watanabe T. 2006. 5' Long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology. 3, 69.
Guo J.U., Su Y., Zhong C., Ming G.L., Song H. 2011. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 145 (3), 423–434.
Hirano S. 2012. Western blot analysis. Methods Mol. Biol. 926, 87–97.
Jiang C., Wu D., Haacke E.M. 2017. Ferritin-EGFP. Chimera as an endogenous dual-reporter for both fluorescence and magnetic resonance imaging in human glioma U251 cells. Tomography. 3 (1), 1–8.
Kumaki Y., Oda M., Okano M. 2008. QUMA: Quantification tool for methylation analysis. Nucleic Acids Res. 36 (Web Server issue), W170–W175.
Power D., Santoso N., Dieringer M., Yu J., Huang H., Simpson S., Seth I., Miao H., Zhu J. 2015. IFI44 suppresses HIV-1 LTR promoter activity and facilitates its latency. Virology. 481, 142–150.
Fouse S.D., Nagarajan R.O., Costello J.F. 2010. Genome-scale DNA methylation analysis. Epigenomics. 2 (1), 105–117.
Du J., Gay M.C.L., Lai C.T., Trengove R.D., Hartmann P.E., Geddes D.T. 2017. Comparison of gravimetric, creamatocrit and esterified fatty acid methods for determination of total fat content in human milk. Food Chem. 217, 505–510.
Euser A.M., Dekker F.W., le Cessie S. 2008. A practical approach to Bland–Altman plots and variation coefficients for log transformed variables. J. Clin. Epidemiol. 61 (10), 978–982.
Thorne N., Inglese J., Auld D.S. 2010. Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem. Biol. 17 (6), 646–657.
Yun C., Dasgupta R. 2014. Luciferase reporter assay in Drosophila and mammalian tissue culture cells. Curr. Protoc. Chem. Biol. 6 (1), 7–23.
Xie W., Silvers R., Ouellette M., Wu Z., Lu Q., Li H., Gallagher K., Johnson K., Montoute M. 2016. A luciferase reporter gene system for high-throughput screening of gamma-globin gene activators. Methods Mol. Biol. 1439, 207–226.
Stables J., Scott S., Brown S., Roelant C., Burns D., Lee M.G., Rees S. 1999. Development of a dual glow-signal firefly and Renilla luciferase assay reagent for the analysis of G-protein coupled receptor signalling. J. Recept. Signal Transduct. Res. 19, 395–410.
Gadkar V., Filion M. 2014. New developments in quantitative real-time polymerase chain reaction technology. Curr. Issues Mol. Biol. 16, 1–6.
Cooksey R.C., Crawford J.T., Jacobs W.R., Jr., Shin-nick T.M. 1993. A rapid method for screening antimicrobial agents for activities against a strain of Mycobacterium tuberculosis expressing firefly luciferase. Antimicrob. Agents Chemother. 37 (6), 1348–1352.
Baldari C.T., Di Somma M.M., Majolini M.B., Ulivieri C., Milia E., Telford J.L. 1998. NF-AT-luciferase reporter T cell lines as tools to screen immunosuppressive drugs. Biol. J. Int. Assoc. Biol. Standard. 26 (1), 1–5.
Michelini E., Cevenini L., Calabretta M.M., Calabria D., Roda A. 2014. Exploiting in vitro and in vivo bioluminescence for the implementation of the three Rs principle (replacement, reduction, and refinement) in drug discovery. Anal. Bioanal. Chem. 406 (23), 5531–5539.
Henderson L.J., Reoma L.B., Kovacs J.A., Nath A. 2020. Advances toward curing HIV-1 infection in tissue reservoirs. J. Virol. 94 (3), e00375–e00319.
Kauder S.E., Bosque A., Lindqvist A., Planelles V., Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathogens. 5 (6), 1–15.
Bouchat S., Delacourt N., Kula A., Darcis G., van Driessche B., Corazza F., Gatot J.S., Melard A., Vanhulle C., Kabeya K., Pardons M., Avettand-Fenoel V., Clumeck N., De Wit S., Rohr O., et al. 2016. Sequential treatment with 5-aza-2'-deoxycytidine and deacetylase inhibitors reactivates HIV-1. EMBO Mol. Med. 8 (2), 117–138.
Greenberg M.V.C., Bourćhis D. 2019. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20 (10), 590–607.
Funding
This research was supported by the National Natural Science Foundation of China (nos. 81501734, 81803308), Natural Science Foundation of Gansu Province, China (no. 20JR5RA215), the Fundamental Research Funds for Central Universities (no. lzujbky-2015-260).
Author information
Authors and Affiliations
Contributions
Xiaoxia Wang and Huaijie Jia conceived and designed the study. Xiaoxia Wang, Huaijie Jia and Xiaole Wei performed the experiments and wrote the paper. Yanrong Lv, Huihui Sun and Jiying Tan analyzed the data. Zhizhong Jing reviewed and edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
This article does not contain any research involving humans or animals as subjects of research.
The authors declare no conflicts of interest.
ADDITIONAL INFORMATION
The text was submitted by the author(s) in English.
Rights and permissions
About this article
Cite this article
Wang, X.X., Jia, H.J., Lv, Y.R. et al. A Luciferase-EGFP Reporter System for the Evaluation of DNA Methylation in Mammalian Cells. Mol Biol 55, 742–751 (2021). https://doi.org/10.1134/S0026893321040099
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893321040099