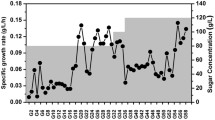
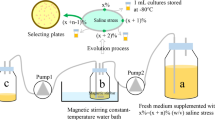
Abstract
High values of agitation and temperature lead to stressful conditions in the fermentations of Lactococcus lactis due to its aero-tolerant and mesophilic nature. Here, the adaptive laboratory evolution (ALE) technique was applied to increase biomass and nisin production yields by enhancing L. lactis subsp. lactis robustness at higher growth temperature and aeration rates. In two separate ALE experiments, after 162 serial transfers, optimum agitation and growth temperature of L. lactis were shifted from 40 rpm and 30 °C to 200 rpm and 37 °C, respectively. Oxidative and acid resistance were enhanced in the evolved strain. Whole-genome sequencing revealed the emergence of five single-nucleotide polymorphisms in the genome of the evolved strain in jag, DnaB, ArgR, cation transporter genes, and one putative protein. The evolved strain of L. lactis in this study has more industrial desirable features and improved nisin production capability and can act more efficiently in nisin production in stressful conditions.
Graphical abstract









Similar content being viewed by others
Availability of Data and Materials
The genomic sequence data were deposited and available in GenBank under the accession number SAMN12990279. The bacterial strains were deposited in the University of Tehran Microorganisms Collection.
References
Özel, B., Şimşek, Ö., Akçelik, M., & Saris, P. E. (2018). Innovative approaches to nisin production. Applied microbiology and biotechnology, 102(15), 6299–6307.
Shin, J. M., Gwak, J. W., Kamarajan, P., Fenno, J. C., Rickard, A. H., & Kapila, Y. L. (2016). Biomedical applications of nisin. Journal of applied microbiology, 120(6), 1449–1465.
Song, A. A.-L., In, L. L., Lim, S. H. E., & Rahim, R. A. (2017). A review on Lactococcus lactis: From food to factory. Microbial cell factories, 16(1), 1–15.
Peterbauer, C., Maischberger, T., & Haltrich, D. (2011). Food-grade gene expression in lactic acid bacteria. Biotechnology journal, 6(9), 1147–1161.
Azizpour, M., Hosseini, S., Jafari, P., & Akbary, N. (2016). Lactococcus lactis as a live delivery vector. Vaccine Research, 3(3), 39–43.
Abbasiliasi, S., Tan, J. S., Ibrahim, T. A. T., Bashokouh, F., Ramakrishnan, N. R., Mustafa, S., & Ariff, A. B. (2017). Fermentation factors influencing the production of bacteriocins by lactic acid bacteria: a review. Rsc Advances, 7(47), 29395–29420.
Jiang, L., Liu, Y., Yan, G., Cui, Y., Cheng, Q., Zhang, Z., Meng, Q., Teng, L., & Ren, X. (2015). Aeration and fermentation strategies on nisin production. Biotechnology letters, 37(10), 2039–2045.
Hao, P., Liang, D., Cao, L., Qiao, B., Wu, H., Caiyin, Q., Zhu, H., & Qiao, J. (2017). Promoting acid resistance and nisin yield of Lactococcus lactis F44 by genetically increasing D-Asp amidation level inside cell wall. Applied microbiology and biotechnology, 101(15), 6137–6153.
Dragosits, M., & Mattanovich, D. (2013). Adaptive laboratory evolution–principles and applications for biotechnology. Microbial cell factories, 12(1), 1–17.
Papadimitriou, K., Alegría, Á., Bron, P. A., De Angelis, M., Gobbetti, M., Kleerebezem, M., Lemos, J. A., Linares, D. M., Ross, P., & Stanton, C. (2016). Stress physiology of lactic acid bacteria. Microbiology and Molecular Biology Reviews, 80(3), 837–890.
Chen, J., Shen, J., Hellgren, L. I., Jensen, P. R., & Solem, C. (2015). Adaptation of Lactococcus lactis to high growth temperature leads to a dramatic increase in acidification rate. Scientific reports, 5(1), 1–15.
Smith, W. M., Pham, T. H., Lei, L., Dou, J., Soomro, A. H., Beatson, S. A., Dykes, G. A., & Turner, M. S. (2012). Heat resistance and salt hypersensitivity in Lactococcus lactis due to spontaneous mutation of llmg_1816 (gdpP) induced by high-temperature growth. Applied and environmental microbiology, 78(21), 7753–7759.
López-González, M. J., Campelo, A. B., Picon, A., Rodríguez, A., & Martínez, B. (2018). Resistance to bacteriocin Lcn972 improves oxygen tolerance of Lactococcus lactis IPLA947 without compromising its performance as a dairy starter. BMC microbiology, 18(1), 1–10.
Şimşek, Ö., Akkoç, N., Con, A., Özçelik, F., Saris, P., & Akçelik, M. (2009). Continuous nisin production with bioengineered Lactococcus lactis strains. Journal of Industrial Microbiology and Biotechnology, 36(6), 863–871.
Lv, W., Cong, W., & Cai, Z. (2004). Nisin production by Lactococcus lactis subsp. lactis under nutritional limitation in fed-batch culture. Biotechnology letters, 26(3), 235–238.
Ariana, M., & Hamedi, J. (2017). Enhanced production of nisin by co-culture of Lactococcus lactis sub sp. lactis and Yarrowia lipolytica in molasses based medium. Journal of biotechnology, 256, 21–26.
Sakthiselvan, P., Meenambiga, S.S., & Madhumathi, R. (2019). Kinetic Studies on Cell Growth, in Cell Growth. IntechOpen.
Pongtharangkul, T., & Demirci, A. (2004). Evaluation of agar diffusion bioassay for nisin quantification. Applied microbiology and biotechnology, 65(3), 268–272.
Burchell, M. J., Mann, J., & Bunch, A. W. (2004). Survival of bacteria and spores under extreme shock pressures. Monthly Notices of the Royal Astronomical Society, 352(4), 1273–1278.
Pericone, C. D., Park, S., Imlay, J. A., & Weiser, J. N. (2003). Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. Journal of bacteriology, 185(23), 6815–6825.
Kim, W. S., Ren, J., & Dunn, N. W. (1999). Differentiation of Lactococcus lactis subspecies lactis and subspecies cremoris strains by their adaptive response to stresses. FEMS Microbiology Letters, 171(1), 57–65.
Shehata, M. G., El-Sahn, M. A., El Sohaimy, S. A., & Youssef, M. M. (2019). In vitro assessment of hypocholesterolemic activity of Lactococcus lactis subsp. lactis. Bulletin of the National Research Centre, 43(1), 60.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., Lesin, V. M., Nikolenko, S. I., Pham, S., & Prjibelski, A. D. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of computational biology, 19(5), 455–477.
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., Formsma, K., Gerdes, S., Glass, E. M., & Kubal, M. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC genomics, 9(1), 1–15.
Lowe, T. M., & Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic acids research, 44(W1), W54–W57.
Lagesen, K., Hallin, P., Rødland, E. A., Stærfeldt, H.-H., Rognes, T., & Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic acids research, 35(9), 3100–3108.
Seemann. (2015). snippy: fast bacterial variant calling from NGS reads. https://githubcom/tseemann/snippy.
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., Land, S. J., Lu, X., & Ruden, D. M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6(2), 80–92.
Smith, W. M., Dykes, G. A., Soomro, A. H., & Turner, M. S. (2010). Molecular mechanisms of stress resistance in Lactococcus lactis. Topics in Applied Microbiology and Microbial Biotechnology, 1106–1118.
Oliveira, L. C., Saraiva, T. D., Silva, W. M., Pereira, U. P., Campos, B. C., Benevides, L. J., Rocha, F. S., Figueiredo, H. C., Azevedo, V., & Soares, S. C. (2017). Analyses of the probiotic property and stress resistance-related genes of Lactococcus lactis subsp. lactis NCDO 2118 through comparative genomics and in vitro assays. PLoS One, 12(4), e0175116.
Zhang, C., Wohlhueter, R., & Zhang, H. (2016). Genetically modified foods: A critical review of their promise and problems. Food Science and Human Wellness, 5(3), 116–123.
Serrazanetti, D. I., Gottardi, D., Montanari, C., & Gianotti, A. (2013). Dynamic stresses of lactic acid bacteria associated to fermentation processes, in Lactic Acid Bacteria-R & D for Food. Health and Livestock Purposes.
Mall, P., Mohanty, B. K., Patankar, D. B., Mody, R., & Tunga, R. (2010). Physiochemical parameters optimization for enhanced nisin production by Lactococcus lactis (MTCC 440). Brazilian Archives of Biology and Technology, 53(1), 203–209.
Dussault, D., Vu, K. D., & Lacroix, M. (2016). Enhancement of nisin production by Lactococcus lactis subsp. lactis. Probiotics and antimicrobial proteins, 8(3), 170–175.
Waites, M. J., Morgan, N. L., Rockey, J. S., & Higton, G. (2009). Industrial microbiology: An introduction. John Wiley & Sons.
Baltz, R. H., Demain, A. L., & Davies, J. E. (2010). Manual of industrial microbiology and biotechnology. American Society for Microbiology Press.
De Vuyst, L., & Vandamme, E. J. (1992). Influence of the carbon source on nisin production in Lactococcus lactis subsp. lactis batch fermentations. Microbiology, 138(3), 571–578.
Kim, W., Hall, R., & Dunn, N. (1997). The effect of nisin concentration and nutrient depletion on nisin production of Lactococcus lactis. Applied Microbiology and Biotechnology, 48(4), 449–453.
Fenster, K., Freeburg, B., Hollard, C., Wong, C., Rønhave Laursen, R., & Ouwehand, A. C. (2019). The production and delivery of probiotics: A review of a practical approach. Microorganisms, 7(3), 83.
Sun, X., Ge, F., Xiao, C.-L., Yin, X.-F., Ge, R., Zhang, L.-H., & He, Q.-Y. (2010). Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. Journal of proteome research, 9(1), 275–282.
Ulrych, A., Holečková, N., Goldová, J., Doubravová, L., Benada, O., Kofroňová, O., Halada, P., & Branny, P. (2016). Characterization of pneumococcal Ser/Thr protein phosphatase phpP mutant and identification of a novel PhpP substrate, putative RNA binding protein Jag. BMC microbiology, 16(1), 1–19.
Frees, D., Vogensen, F. K., & Ingmer, H. (2003). Identification of proteins induced at low pH in Lactococcus lactis. International journal of food microbiology, 87(3), 293–300.
Rallu, F., Gruss, A., Ehrlich, S. D., & Maguin, E. (2000). Acid- and multistress-resistant mutants of Lactococcus lactis: Identification of intracellular stress signals. Molecular microbiology, 35(3), 517–528.
Haney, C. J., Grass, G., Franke, S., & Rensing, C. (2005). New developments in the understanding of the cation diffusion facilitator family. Journal of Industrial Microbiology and Biotechnology, 32(6), 215–226.
Turner, M. S., Tan, Y. P., & Giffard, P. M. (2007). Inactivation of an iron transporter in Lactococcus lactis results in resistance to tellurite and oxidative stress. Applied and Environmental Microbiology, 73(19), 6144–6149.
Larsen, R., Buist, G., Kuipers, O. P., & Kok, J. (2004). ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. Journal of bacteriology, 186(4), 1147–1157.
Xiong, L., Teng, J. L., Watt, R. M., Liu, C., Lau, S. K., & Woo, P. C. (2015). Molecular characterization of arginine deiminase pathway in Laribacter hongkongensis and unique regulation of arginine catabolism and anabolism by multiple environmental stresses. Environmental microbiology, 17(11), 4469–4483.
Cheng, C., Dong, Z., Han, X., Sun, J., Wang, H., Jiang, L., Yang, Y., Ma, T., Chen, Z., & Yu, J. (2017). Listeria monocytogenes 10403S arginine repressor ArgR finely tunes arginine metabolism regulation under acidic conditions. Frontiers in microbiology, 8, 145.
Budin-Verneuil, A., Maguin, E., Auffray, Y., Ehrlich, S. D., & Pichereau, V. (2004). An essential role for arginine catabolism in the acid tolerance of Lactococcus lactis MG1363. Le Lait, 84(1-2), 61–68.
Huang, R., Pan, M., Wan, C., Shah, N. P., Tao, X., & Wei, H. (2016). Physiological and transcriptional responses and cross protection of Lactobacillus plantarum ZDY2013 under acid stress. Journal of dairy science, 99(2), 1002–1010.
Wu, H., Liu, J., Miao, S., Zhao, Y., Zhu, H., Qiao, M., Saris, P. E. J., & Qiao, J. (2018). Contribution of YthA, a PspC family transcriptional regulator of Lactococcus lactis F44 acid tolerance and nisin yield: A transcriptomic approach. Applied and environmental microbiology, 84(6).
Li, Y., & Araki, H. (2013). Loading and activation of DNA replicative helicases: The key step of initiation of DNA replication. Genes to Cells, 18(4), 266–277.
Saluja, D., & Godson, G. N. (1995). Biochemical characterization of Escherichia coli temperature-sensitive dnaB mutants dnaB8, dnaB252, dnaB70, dnaB43, and dnaB454. Journal of Bacteriology, 177(4), 1104–1111.
Carney, S. M., Gomathinayagam, S., Leuba, S. H., & Trakselis, M. A. (2017). Bacterial DnaB helicase interacts with the excluded strand to regulate unwinding. Journal of Biological Chemistry, 292(46), 19001–19012.
Naranjo-Briceño, L., Pernía, B., Guerra, M., Demey, J. R., De Sisto, Á., Inojosa, Y., González, M., Fusella, E., Freites, M., & Yegres, F. (2013). Potential role of oxidative exoenzymes of the extremophilic fungus Pestalotiopsis palmarum BM-04 in biotransformation of extra-heavy crude oil. Microbial biotechnology, 6(6), 720–730.
Nitharwal, R. G., Paul, S., Dar, A., Choudhury, N. R., Soni, R. K., Prusty, D., Sinha, S., Kashav, T., Mukhopadhyay, G., & Chaudhuri, T. K. (2007). The domain structure of Helicobacter pylori DnaB helicase: The N-terminal domain can be dispensable for helicase activity whereas the extreme C-terminal region is essential for its function. Nucleic acids research, 35(9), 2861–2874.
Author information
Authors and Affiliations
Contributions
RP has done the experiments and wrote the manuscript, and JH designed and supervised the experiments and edited the manuscript.
Corresponding author
Ethics declarations
Ethical Approval
The authors, Reyhaneh Papiran (RP) and Javad Hamedi (JH), considered all related ethical and bioethical rules.
Consent to Participate
The authors consented to participate the current research.
Consent for Publication
The authors approved the manuscript and agree to publish it in current form in ABAB.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Papiran, R., Hamedi, J. Adaptive Evolution of Lactococcus Lactis to Thermal and Oxidative Stress Increases Biomass and Nisin Production. Appl Biochem Biotechnol 193, 3425–3441 (2021). https://doi.org/10.1007/s12010-021-03609-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03609-6