Spatial Scale Dependence of Ecological Factors That Regulate Functional and Phylogenetic Assembly in a Mediterranean High Mountain Grassland
- 1Departamento de Farmacología, Farmacognosia y Botánica, Facultad de Farmacia, Universidad Complutense de Madrid, Madrid, Spain
- 2Área de Biodiversidad y Conservación, Departamento de Biología y Geología, Física y Química Inorgánica, Escuela Superior de Ciencias Experimentales y Tecnológicas, Universidad Rey Juan Carlos, Móstoles, Spain
- 3Centro de Investigaciones Sobre Desertificación (CSIC-UV-GV), Valencia, Spain
- 4Department of Environmental Systems Science, Swiss Federal Institute of Technology Zurich (ETH), Zurich, Switzerland
- 5Museo Nacional de Ciencias Naturales MNCN (CSIC), Madrid, Spain
Understanding how functional and phylogenetic patterns vary among scales and along ecological gradients within a given species pool is critical for inferring community assembly processes. However, we lack a clear understanding of these patterns in stressful habitats such as Mediterranean high mountains where ongoing global warming is expected to affect species fitness and species interactions, and subsequently species turnover. In this study, we investigated 39 grasslands with the same type of plant community and very little species turnover across an elevation gradient above the treeline at Sierra de Guadarrama National Park in central Spain. In particular, we assessed functional and phylogenetic patterns, including functional heterogeneity, using a multi-scale approach (cells, subplots, and plots) and determined the relevance of key ecological factors (i.e., elevation, potential solar radiation, pH, soil organic carbon, species richness, and functional heterogeneity) that affect functional and phylogenetic patterns at each spatial scale. Overall, at the plot scale, coexisting species tended to be more functionally and phylogenetically similar. By contrast, at the subplot and cell scales, species tended to be more functionally different but phylogenetically similar. Functional heterogeneity at the cell scale was comparable to the variation across plots along the gradient. The relevance of ecological factors that regulate diversity patterns varied among spatial scales. An increase in elevation resulted in functional clustering at larger scales and phylogenetic overdispersion at a smaller scale. The soil pH and organic carbon levels exhibited complex functional patterns, especially at small spatial scales, where an increase in pH led to clustering patterns for the traits related to the leaf economic spectrum (i.e., foliar thickness, specific leaf area, and leaf dry matter content). Our findings confirm the presence of primary environmental filters (coldness and summer drought at our study sites) that constrain the regional species pool, suggesting the presence of additional assembly mechanisms that act at the smallest scale (e.g., micro-environmental gradients and/or species interactions). Functional and phylogenetic relatedness should be determined using a multi-scale approach to help interpret community assembly processes and understand the initial community responses to environmental changes, including ongoing global warming.
Introduction
Coexistence theory considers that community assembly is a process regulated by multiple mechanisms, both stochastic and deterministic, which act at different spatial scales and that are subject to different ecological factors (Palmer, 1994; Chesson, 2000). Understanding how these assembly mechanisms and their drivers operate simultaneously at different spatial scales and along environmental gradients is critical for forecasting community responses to environmental changes, including the major issue of ongoing global warming. Approaches based on functional traits, which are valuable indicators of resource use strategies and fitness, can provide critical information about how ecological factors affect assembly because only species with the appropriate set of traits can be part of the local (or realized) plant community (Cornelissen et al., 2003; Violle et al., 2007). In addition, phylogeny seems to be involved in assembly processes because it summarizes the evolutionary history of the local species pool (Losos, 1996; Webb et al., 2002; Cavender-Bares et al., 2009; Kraft and Ackerly, 2010; Mouquet et al., 2012; Gerhold et al., 2015), and it is often used as a proxy of current functional information as it is assumed that evolutionary diversification has generated predictable trait diversification (Flynn et al., 2011). Thus, comparisons of the functional (and/or phylogenetic) diversity of species assemblages and those expected from null assemblages have been performed routinely during the last two decades to identify the assembly processes that structure plant communities at different spatial scales (Díaz et al., 1998; Cavender-Bares et al., 2006; Kraft and Ackerly, 2010; Chalmandrier et al., 2017; López-Angulo et al., 2021).
Among the so-called deterministic processes, environmental filtering and limiting similarity are two major ecological forces that drive community assembly. It is known that these mechanisms act at different spatial scales on the regional species pool (Levin, 1992; Swenson et al., 2006; Emerson and Gillespie, 2008; Götzenberger et al., 2012) but their importance in specific plant communities has traditionally been studied at a single spatial scale under similar environmental conditions (but see Kraft and Ackerly, 2010; Messier et al., 2010; de Bello et al., 2013b; Carboni et al., 2014; Chalmandrier et al., 2017) and/or in systems with high species turnover. Environmental filtering, which is considered to act at relatively large scales, is expected to lead to a community where species are more functionally similar or closely related (i.e., functional and phylogenetic clustering; Weiher and Keddy, 1995; Leibold, 1998; Webb, 2000) because filters will select species that can thrive under the local conditions (but see Gerhold et al., 2015). By contrast, limiting similarity (sensu MacArthur and Levins, 1967), which is related to the existence of niche differences where plant-to-plant competitive interactions prevail (i.e., smaller scales), predicts that competitiveness among species with similar requirements leads to niche segregation where the coexisting species are functionally and/or phylogenetically different (Mason and Wilson, 2006; Kraft et al., 2008), thereby implying trait and phylogenetic overdispersion for exploiting the diversity of niches at small scales. However, alternative combinations have also been described. For example, strong environmental heterogeneity may potentially lead to a functional overdispersion pattern at relatively small spatial scales because different niches are available (Mayfield and Levine, 2010; de Bello et al., 2013b), which would determine intense turnover. In addition, trait clustering may be linked to biotic competitive processes at small spatial scales when the dominance of closely related species excludes weaker competitors (Chesson, 2000; Mayfield and Levine, 2010).
Evaluating patterns of site-to-site functional variation, i.e., a type of β-diversity referred to as functional heterogeneity in the following, among spatial scales may be necessary for understanding the importance of mechanisms involved at both local and regional scales in community assembly (Seabloom et al., 2005; Siefert et al., 2013; Yang et al., 2015). Thus, high functional heterogeneity would be expected when comparing the functional variation between sites or assemblages (at large spatial scales) simply because geographic and environmental distances often imply strong species turnover (i.e., different community types) or shifts in their functional traits (Myers et al., 2013). However, functional heterogeneity could be stabilized at the regional scale when a strong environmental filter acts and selects species with a particular combination of associated functional traits (Körner, 2003). In addition, functional heterogeneity could be increased further at local scales (within sites) by small-scale environmental variations such as those induced by soil heterogeneity (Freestone and Inouye, 2006) or species interactions (Pescador et al., 2014).
Mountains exhibit steeper environmental gradients over short geographic distances and at various scales (Körner, 2003). Thus, mountains are excellent systems for investigating how assembly processes operate at different spatial scales and the relevance of ecological factors for structuring plant communities (McCain and Colwell, 2011). Previous trait- and phylogenetic-based studies in high mountain communities suggest that environmental filtering increases along the elevation by affecting functional and phylogenetic patterns (de Bello et al., 2013a; Pistón et al., 2016; Xu et al., 2017; Le Bagousse-Pinguet et al., 2018). In addition, these patterns can be shaped by local factors in alpine communities, such as the potential solar radiation, soil pH, or nutrient heterogeneity (Cadotte et al., 2011; Bernard-Verdier et al., 2012; Luo et al., 2016). However, functional and phylogenetic patterns have generally been assessed across large environmental gradients, which imply high species turnover with major changes in the habitat type, and thus it is difficult to understand assembly mechanisms at different scales.
In the present study, we investigated functional and phylogenetic patterns as well as their ecological predictors by considering a nested spatial design in a single plant community type located along a 500-m elevation gradient in a Mediterranean high mountain area. This community is characterized by strong environmental filtering acting along the whole study gradient, which excludes species that are vulnerable to cold (Pescador et al., 2016), and a severe summer drought, especially at lower elevations (Giménez-Benavides et al., 2007; Olano et al., 2013). These conditions limit the species pool to specialists that are well adapted to such conditions, thereby resulting in the same type of plant community with low species turnover (Pescador et al., 2015). To the best of our knowledge, a low turnover scenario has not been considered in previous studies for assessing functional and phylogenetic patterns, but it probably represents the best system for investigating how a habitat-specific species pool is affected by ongoing global warming before a change in turnover. Thus, we hypothesized that an environmental adjustment would prevail in this community with environmental filtering acting at large scales and across the elevation gradient, which would define the realized assemblages through functional and phylogenetic clustering, especially at higher elevations. By contrast, micro-environmental heterogeneity and species interactions would prevail at smaller scales, thereby leading to functional and phylogenetic divergence, which should be correlated with the soil characteristics at these scales. As a consequence, the functional heterogeneity should be relatively similar across the spatial scales studied because the primary environmental filters should stabilize the large functional heterogeneity between sites (regional scale), whereas micro-environmental heterogeneity and species interactions could force the opposite. Thus, in the present study, we addressed the following three questions: (i) Are the functional and phylogenetic clustering–overdispersion patterns within assemblages similar across spatial scales? (ii) Does the functional heterogeneity between assemblages vary among spatial scales? (iii) How do functional and phylogenetic patterns vary along the elevation gradient and with other key ecological factors (e.g., potential solar radiation, soil pH or nutrient heterogeneity, and species richness) at each spatial scale?
Materials and Methods
Study Site and Field Sampling
This study was conducted above the treeline at Sierra de Guadarrama National Park located northwest of Madrid in Spain (40°46′39′′ to 40°51′8″ N; 3°49′44″ to 4°4′59″ W; 1,940 to 2,419 m a.s.l.). The climate is Mediterranean, with a mean annual temperature and precipitation of 6.4°C and 1,350 mm, respectively, and a drought from May to October (less than 10% of the annual precipitation), which is more intense at lower elevations (Giménez-Benavides et al., 2007; Olano et al., 2013).
The treeline is located between 1,900 and 2,000 m a.s.l., and it is dominated by Scots pine (Pinus sylvestris L.) interspersed in a shrubby matrix of Cytisus oromediterraneus Rivas Mart. et al. and Juniperus communis L. subsp. alpina (Suter) Čelak. Above this elevation, the dominant shrubby matrix extends up to 2,100–2,200 m a.s.l., with dry and cryophilic grassland dominated by Festuca curvifolia Lag. ex Lange in less steep locations. In the highest areas (from 2,200 to 2,419 m a.s.l.), grassland represents the dominant vegetation and it is structured in a mosaic of patches dominated by F. curvifolia and some creeping and cushion chamaephytes clumped in a bare ground matrix (Pescador et al., 2014).
Thirty-nine sites covering the whole elevation range of F. curvifolia grasslands were selected in the summer of 2011 (Supplementary Figure 1A). At each site, we established a sampling plot of 20 m × 20 m and surveyed the vegetation at three different spatial scales: cell, subplot, and plot (Supplementary Figure 1B). First, a square measuring 2.4 m × 2.4 m was haphazardly located in the center of the plot and divided into 64 cells of 30 cm × 30 cm, which represented the cell scale (smallest scale). Second, five subplots of 2.4 m × 2.4 m (including the grid of cells) were established in the center and four corners of the plot as the intermediate scale. The coverage was visually estimated for every plant species, soil, and rock at the three spatial scales. At the plot scale, all plant species and their percentage cover were estimated as the average cover per species in the five subplots after also considering the presence and cover of the plant species not found in the cells or subplots but present in the plot (for more details, see López-Angulo et al., 2019).
Each plot was also characterized based on a set of environmental variables. In particular, we measured the elevation and orientation using a GPS system (Garmin Colorado-300) and the slope with a clinometer (Silva Clinomaster CM-360-%, LA). Orientation and slope values were employed to estimate the potential solar radiation according to Gandullo’s method (Gandullo, 1974; López-Angulo et al., 2018). Finally, 15 soil samples (diameter = 5 cm, depth = 10 cm) were randomly collected from each plot (Supplementary Figure 1B) under vegetated patches of the dominant grass (usually F. curvifolia; five samples), shrub canopies (C. oromediterraneus and J. communis subsp. alpina; five samples), and on bare ground (five samples). The soil samples were sieved (2-mm mesh) and air dried for 1 month, before the pH and soil organic carbon (SOC, %) were determined at the NutriLab-URJC laboratory. The pH was determined using a pH meter GLP 21 (Crison, Barcelona, Spain) and SOC was determined by colorimetry after oxidation with K2Cr2O2 and H2SO4.
Plant Functional Traits
During the summer of 2011 and 2012, five plant functional traits were measured in a total of 56 species (Supplementary Table 1). These species represented more than 83% of the species in the community study and approximately 99% of the species cover. At least 10 individuals per species were selected and five functional traits were measured according to standardized protocols (Cornelissen et al., 2003): plant height (Hmax; distance between the ground and top of photosynthetic tissues), foliar thickness (FT), specific leaf area (SLA; ratio of leaf fresh area relative to dry weight), leaf dry matter content (LDMC; ratio of leaf dry weight relative to fresh weight), and seed mass (for more details, see Pescador et al., 2015).
DNA Isolation, Sequencing, and Phylogenetic Analysis
We collected fresh young leaves from three individuals per species and they were stored under dry conditions in silica gel for 1 month. Up to 20 mg of each dry leaf sample was disrupted and homogenized with a TissueLyser LT (Qiagen, Valencia, CA, United States) using glass beads. Total genomic DNA was extracted with a DNeasy Plant Mini-Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s instructions. Standard polymerase chain reactions (PCRs) were conducted to amplify two barcoding loci (i.e., rbcL and matK). In the PCR amplification mixture, 2 μl of DNA was added to 23 μl of reaction mixture comprising 2.5 μl of Taq buffer 2 mM with MgCl2, 1 μl of dNTP Mix (0.4 mM), 1.25 μl of reverse and forward primers, and 1.25 U Taq DNA Polymerase (Biotools, Madrid, Spain). PCR was performed with an S1000 Thermal Cycler (Bio-Rad, United States) using the specific PCR conditions given in Supplementary Table 2. The PCR products were cleaned up using ExoSap-IT® (USB Corporation, Cleveland, OH, United States) and submitted to Macrogen Inc. (Seoul, South Korea) for sequencing in the forward and reverse directions. In total, 288 sequences [three individuals per species × (56 species successfully sequenced with rbcL + 40 species successfully sequenced with matK)] were edited with Sequencher 4.1.4 (Genes Code Corporation), and the consensus sequences in each species were determined from the forward and reverse sequences. The rbcL and matK markers were aligned independently using Mafft online-version 7 (Katoh and Standley, 2013) following the G-INS-I strategy recommended for < 200 sequences with global homology. MacClade 4.06 (Maddison and Maddison, 2003) was used to concatenate primer configurations (rbcL + matK) and to create a combined matrix with missing data when the marker could not be sequenced.
The rbcL + matK combined matrix was used to reconstruct the phylogeny topology and the divergence times for our community using Bayesian inference and the Markov chain Monte Carlo (MCMC) approach in the BEAST 1.6.1 program (Drummond and Rambaut, 2007). We ran three MCMC chains with 50 million generations and sampled one tree and their parameters every 10,000 generations (partition). We estimate the rooted, time-measured phylogeny inferred using an uncorrelated relaxed molecular clock, which assumes that the substitution rates associated with each branch are drawn independently from a single lognormal distribution (Drummond et al., 2006). The substitution model used for each partition was GTR + Γ. The substitution model and estimated frequencies were unlinked among partitions. To calibrate the tree temporally, we constrained the divergence times for the minimum crown age at seven nodes based on the fossil record (Supplementary Table 3). Each constrained node was drawn following a lognormal or exponential prior with different parameters (for more details, see Supplementary Table 3). The Tracer 1.6 program (Rambaut et al., 2014) was used to evaluate the effective sample size for parameters, and we verified that three runs converged on the posterior distribution and reached stationarity. We combined the independent MCMC parameters and sample trees using the LogCombiner 1.6.1 program. Finally, all trees were summarized in a single maximum clade credibility tree (Supplementary Figure 2) after removing 2,500 trees as the burn-in and calculating the clade posterior probabilities and 95% honest posterior density intervals for node-specific parameters using TreeAnnotator 1.6.1.
Community Structure Metrics
We calculated the mean pairwise distance (MPDobs; see de Bello et al., 2016) between the coexisting species observed in each cell, subplot, or plot to assess the functional and phylogenetic clustering–overdispersion patterns at different spatial scales. The functional and phylogenetic distance between each pair of coexisting species were calculated based on Gower’s dissimilarity matrix (Pavoine et al., 2009) using the information for the plant functional traits and the cophenetic correlation for the phylogenetic approach. Each MPDobs value was expressed relative to the MPD simulated (MPDexp) based on a null model to calculate a standardized effect size (SES; Gotelli and McCabe, 2002) for each scale, trait, all traits together, and phylogeny. Choosing an appropriate null model is a critical step when testing the effects of different mechanisms (Perronne et al., 2017), so we designed two “realistic” null models where the species names in each simulated assemblage were the names of the species in the plot to which the cell/subplot belonged (i.e., local pool, NM1), or the species names in all plots (i.e., regional pool, NM2). Thus, at the cell and subplot scales, the species that generated the null assemblage could come from the local or regional pool, whereas only the regional pool was considered at the plot scale. In addition, irrespective of the null model used, the number of species and total cover in each cell, subplot, or plot were kept fixed to control for differences in resource availability among scales and species (Gotelli, 2000; Le Bagousse-Pinguet et al., 2018). The likelihood that one species appeared in null assemblages was directly related to its abundance at the plot or regional scale, and the cover for each species in the null distribution was proportional to its abundance at the plot or regional scale. Based on these constraints, we generated 1,000 null assemblages for each cell (64 cells/plot × 39 plots), subplot (5 subplots/plot × 39 plots), and plot (39 plots), where the MPDexp values were estimated using multi-trait, individual trait, and phylogenetic information, and each null model (i.e., NM1 and NM2). Comparing each MPDobs to the corresponding 1,000 random MPDexp values allowed us to calculate the SES of the MPD as follows:
where MPDobs is the MPD obtained from the data observed at the pertinent working scale and using the functional or phylogenetic distance, MPDexp is the mean value for the 1,000 null MPD distributions, and sd(MPDexp) is the standard deviation value for the 1,000 null MPD distributions. It should be noted that this index is equivalent to the inverse of the net relatedness index used in many phylogenetic approaches (Webb et al., 2002), and positive values indicate that the co-occurring species are more functional or phylogenetically distant than in the null assemblages (overdispersion pattern), whereas negative values denote that species are more closely related (clustering pattern). In total, we calculated SES–MPD values for 64 × 39 cells, 5 × 39 subplots, and 39 plots by considering each plant functional trait individually (i.e., Hmax, FT, SLA, LDMC, and seed mass), all traits together, and phylogenetic distances. At the cell and subplot scales, the null assemblage was built by considering the local (NM1) or regional (NM2) species pool. In each situation, the average SES–MPD value per plot was considered for each of the 64 cells and five subplots.
We estimated the coefficient of variation of the community weighted mean (CV-CWM; ratio of the standard deviation to the mean for average traits weighted by species abundances) for the realized assemblages at each spatial scale (i.e., cell, subplot, and plot) to characterize the functional heterogeneity at three spatial scales. We considered the information for 64 cells per plot to estimate the functional heterogeneity within each subplot (i.e., CV-CWMw) because the summed area of the 64 cells was equivalent to the subplot area. We calculated the between-subplot heterogeneity by considering the five subplots per plot (i.e., CV-CWMb). Finally, a unique CV was obtained for the 39 plots at the regional scale (i.e., CV-CWMr). The latter variation component can be considered as the total variation between plots, and thus of the plant community in study area. All of these functional heterogeneity components were estimated by considering all traits together (in a multi-trait approach) and each trait independently (i.e., Hmax, FT, SLA, LDMC, and seed mass).
Statistical Analyses
We performed t-tests using the SES–MPD values at each spatial scale to determine whether the functional and phylogenetic diversities differed significantly from expectations. In particular, SES–MPD values significantly higher than 0 indicated overdispersion among assemblage species, whereas SES–MPD values lower than zero denoted clustering patterns. We performed paired t-test to evaluate the differences between functional heterogeneity within (CV-CWMw) and between subplots (CV-CWMb). We established linear models to assess the response of ecological factors on each SES–MPD following a multi-model inference (MMI) over all possible combinations of the factors and with AICc. A total of 35 independent linear models were drawn as a result of considering SES–MPD values for all traits together, each individual trait, and phylogeny at each of the three spatial scales and two null models. We used non-correlated ecological factors (i.e., elevation, potential solar radiation, pH, SOC, and species richness). We also considered the functional heterogeneity as an additional predictor factor. Within each independent model, all possible combinations of ecological factors were considered, and the resulting “partial models” were ranked according to the AICc value with respect to the “partial model” with the minimum AICc (ΔAICc = AICi – AICmin). “Partial models” with ΔAICc < 2 were considered to be indistinguishable (Burnham and Anderson, 2002) and an average explanatory model was defined according to this threshold. The average estimate, significance, and relative importance of each predictor (w+) were estimated for each model. The relative importance of a predictor was evaluated by summing the wi values for the partial models that contained the predictor of interest. Finally, the phylogenetic signal was calculated for each functional trait with Blomberg’s K index to capture the effect of trait evolution and to assess the existence of phylogenetic niche conservatism in our system. Basically, Blomberg’s K represents the ratio of the observed phylogenetically correct mean-square error divided by the mean-square error using a variance matrix derived from the phylogeny standardized by the expectation under a Brownian motion model (Blomberg et al., 2003). The significance of K (p-value) was calculated by comparing it to the null distribution with 9,999 replicates. Thus, K < 1 indicates that closely related species resemble each other less than expected under the Brownian motion model of trait evolution (no phylogenetic signal), whereas K > 1 suggests that closely related species are more similar than predicted by the model (stronger phylogenetic signal).
All statistical analyses were conducted with R 3.6.0 (R Core Team, 2019) and the following functions: dbFD and gowdis functions implemented in the FD package (Laliberté and Legendre, 2010; Laliberté and Shipley, 2011), dredge and importance functions in the MuMIn package (Kamil, 2013), melodic function (de Bello et al., 2016), and phylosignal function in the picante package (Kembel et al., 2010).
Results
Overall, we identified significant deviations in the functional and phylogenetic clustering–overdispersion patterns depending on the spatial scale (i.e., cell, subplot, or plot scale) and null model (i.e., species consideration from the plot pool, NM1, or from the regional pool, NM2; Figure 1 and Supplementary Table 4). At the plot scale, the trait clustering pattern (SES–MPD values < 0) was most relevant for all single-trait and multi-trait approaches. At the subplot and cell spatial scales, significant trait overdispersion (SES–MPD values > 0) had considerable importance for most traits irrespective of whether we considered the plot (NM1) or regional pool species (NM2). By contrast, Hmax had a significant clustering pattern at both spatial scales and when considering both null models (Figure 1B). The community had a weak phylogenetic clustering pattern at the plot scale (species at plot scales were more closely related than expected from the null model), and significant clustering was found at the subplot and cell scales when both null models were used (Figure 1G).
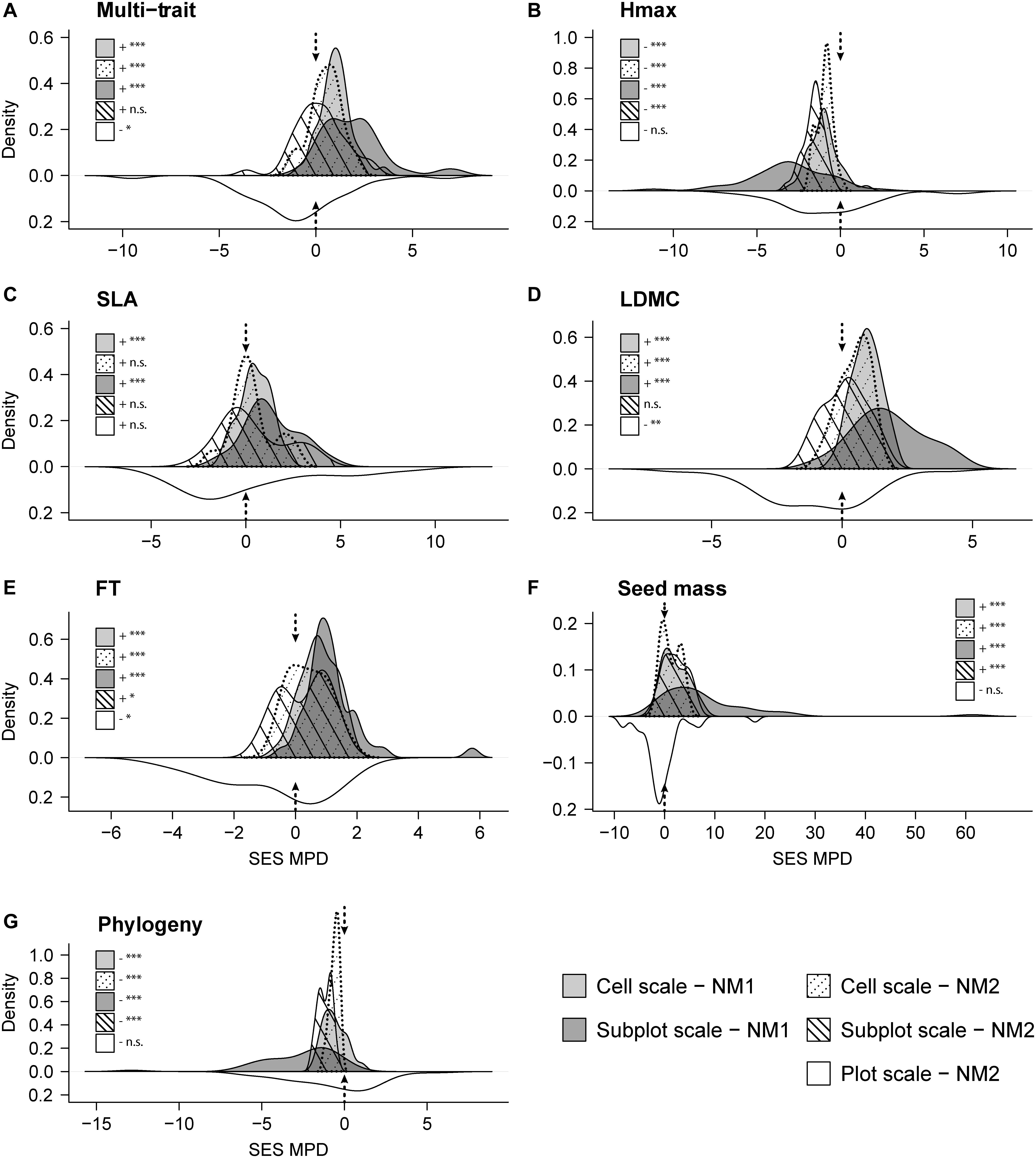
Figure 1. Density plots of standardized effect size of mean pairwise distance (SES–MPD) for (A) multi-trait, (B–F) each individual plant functional trait and (G) phylogeny. Three different operating spatial scales (cell, subplot, or plot scale) and two null models (simulated assemblages using plot pool, NM1, and regional pool, NM2) are included in each plot. Negative values indicate that co-occurring species within the assemblages are more related than expected by chance (i.e., clustering), whereas positive values denote distances greater than expected by chance (i.e., overdispersion). Black-dashed arrows represent the SES threshold of 0. Each panel shows the results of one-sample t-tests conducted to determine whether SES–MPD values for each approach, null model, and scale were positive (+) or negative (–), and significantly different from 0 as: ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05. Full scores for the one-sample t-tests are presented in Supplementary Table 4. Hmax, plant height; SLA, specific leaf area; LDMC, leaf dry matter content; FT, foliar thickness.
The functional heterogeneity across spatial scales exhibited similar patterns for the multi-trait approach and based on each plant functional trait (Figure 2 and Supplementary Table 5). On average, the functional heterogeneity was usually greater at the regional scale (i.e., CV-CWMr) than those within and between subplots (i.e., CV-CWMw and CV-CWMb, respectively), except for traits related to the leaf economic spectrum (i.e., FT, SLA, and LDMC), where the values of CV-CWMw were similar or even higher than those of CV-CWMr. Paired t-tests between the CVs at the plot level (i.e., CV-CWMw and CV-CWMb) indicated lower variability between subplots than within subplots in most cases, except for Hmax (Figure 2B) and FT (Figure 2C), where both components were not significantly different (p-values according to paired t-test = 0.50 and 0.09, respectively; Supplementary Table 5).

Figure 2. Relationships among coefficient of variation (CV) of community weighted mean (CWM) at the three spatial scales assessed [within subplots (CV-CWMw; y-axis in each panel), between subplots (CV-CWMb; x-axis in each panel), and between plots (CV-CWMr; solid lines in each panel)] and considering (A) multi-trait approach and (B–F) each individual plant functional trait. Dashed lines represent a 1:1 relationship. In each panel, p-values obtained from paired t-tests for within subplots (w), between subplots (b), and between plots (r) are denoted as: ***p < 0.001; **p < 0.01; *p < 0.05. Full results for these paired t-tests are presented in Supplementary Table 5. Hmax, plant height; FT, foliar thickness; SLA, specific leaf area; LDMC, leaf dry matter content.
The effects of ecological factors on the SES–MPD values depended on the spatial scale and the null model considered (Figure 3). In particular, clustering patterns (negative relationships) were detected for multi-trait, SLA, and seed mass as the elevation increased at the plot scale and under NM2. By contrast, a phylogenetic overdispersion pattern (positive relationship) was more obvious at the smallest spatial scale and under NM2 as the elevation increased. The potential solar radiation effect led to clustering patterns for multi-trait and seed mass for co-existing species at the subplot scale. Among the soil factors, an increase in SOC resulted in the overdispersion of SLA at different scales but a clustering pattern of FT at the plot and subplot scales and under NM2. Higher soil pH levels resulted in overdispersion patterns for Hmax and seed mass at the subplot and cell scale, respectively. By contrast, clustering patterns were found for FT, SLA, and LDMC when the soil pH levels increased at the subplot scale and under NM1. Increased species richness was related to clustering patterns for multi-trait, Hmax, FT, and phylogeny at the three spatial scales. Finally, overdispersion patterns for Hmax and FT at the cell scale were directly dependent on the functional heterogeneity characterized as the CV of the corresponding CWM.
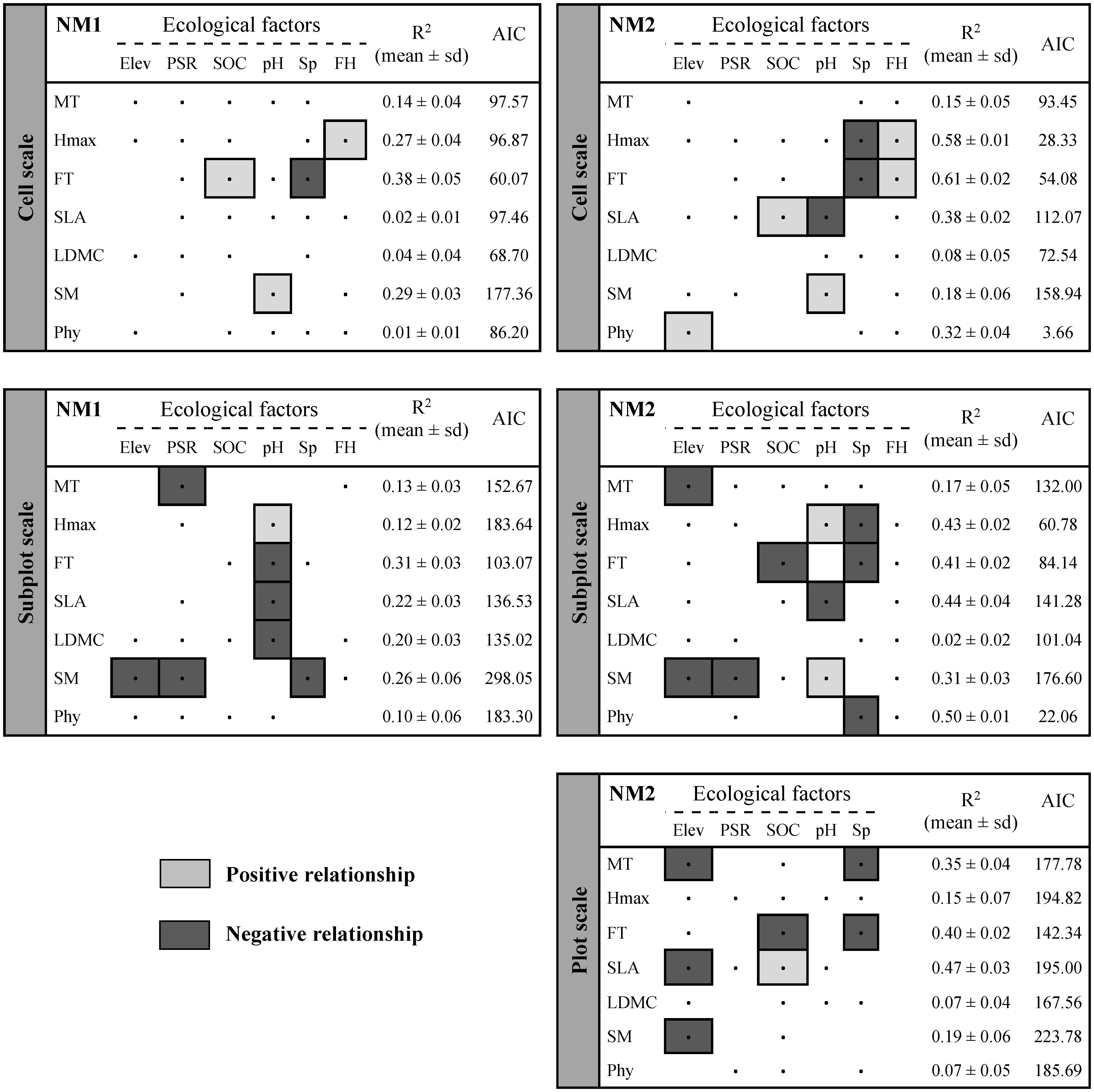
Figure 3. Linear averaged models showing the responses of non-correlated ecological factors on each standardized effect size of mean pairwise distance (SES–MPD) for multi-trait, individual functional traits, and phylogenetic approaches. SES–MPD values were considered at three spatial scales (i.e., cell, subplot, or plot scale) and under two null models (i.e., simulated assemblages using plot pool, NM1, and regional pool, NM2). In each case, all possible predictor combinations were tested and “partial models” were ranked according to Akaike’s information criterion (AICc). Averaged models were drawn from the best partial models based on ΔAICc < 2 (ΔAICc = AICi – AICmin). Predictors included in each averaged model are denoted by a dot (.) and positive or negative significant relationships (according to the average-slope value) are highlighted in light gray or dark gray, respectively. R2 (mean ± standard deviation) and AICc values are shown in each case for the best partial models. Full results for the averaged linear models are shown in Supplementary Table 6. MT, multi-trait; Hmax, plant height; FT, foliar thickness; SLA, specific leaf area; LDMC, leaf dry matter content; SM, seed mass; Phy, phylogeny; Elev, elevation (m a.s.l.); PSR, potential solar radiation; SOC, soil organic carbon (%); pH, soil pH; Sp, number of species per plot; FH, functional heterogeneity (CV-CWM).
Discussion
Our results indicate the existence of three assembly mechanisms operating at different spatial scales in the same plant community type on a Mediterranean high mountain. First, we established the scale dependence of the mechanisms involved in the assembly process ranging from functional clustering at regional spatial scales to functional overdispersion at the smallest scale (Mason et al., 2011; de Bello et al., 2013b; Carboni et al., 2014), and phylogenetic clustering irrespective of the spatial scale. Second, we found that the functional heterogeneity at the smallest scale represented a source of variation, which was as important as the heterogeneity detected along the whole elevation gradient, especially for the traits related to the leaf economic spectrum. Third, we showed that different ecological factors (i.e., elevation, potential solar radiation, SOC, pH, species richness, and functional heterogeneity) regulated the functional and phylogenetic patterns among co-occurring species and ultimately determined the community assembly. Overall, the results obtained in our study confirmed the importance of considering approaches based on functional traits and phylogenetic information across spatial scales in plant coexistence assessments (Kraft and Ackerly, 2010; Perronne et al., 2014).
Diversity Patterns Across Spatial Scales
By assessing the SES–MPD according to two null models (simulated community assemblages using plot pool, NM1, and regional pool, NM2) and at three spatial scales (cell, subplot, or plot scale), we detected contrasting clustering–overdispersion patterns in the functional and phylogenetic diversity (Freschet et al., 2011; Bernard-Verdier et al., 2012; de Bello et al., 2013b). We found that an increase in the spatial granularity (i.e., plot scale of 20 m × 20 m) implied a clustering pattern for most plant functional traits, thereby supporting the idea that the effects of environmental filters are more conspicuous at larger scales (Weiher and Keddy, 1995). By contrast, when the spatial granularity was reduced (i.e., subplot and cell scales), functional overdispersion was the most frequent pattern for all functional traits, except for Hmax. Similarly, some previous studies found an overdispersion pattern at small spatial scales (Stubbs and Wilson, 2004; de Bello et al., 2013b), and our results support the limiting similarity hypothesis for most functional traits at these scales. Hmax was the only functional trait that exhibited a clustering pattern at these scales (Figure 1B), and thus the heights of species in a subplot or cell tended to be similar. In high mountain areas, cold is a strong abiotic filter and greater height may be considered a limitation for the survival of species (Körner, 2003). Consequently, low stature appears to be the most suitable strategy for this type of environment. Furthermore, Mayfield and Levine (2010) suggested the existence of similarity patterns in Hmax linked to competitive ability. The phylogenetic relationship assessment detected clustering patterns at three study scales, thereby indicating that the species were more closely related than the null expectation. This pattern is consistent with the prevalence of environmental filtering indicated for most of our functional traits and the idea that phylogenetic relatedness suggests functional proximity (Webb, 2000; Cavender-Bares et al., 2006; Kraft and Ackerly, 2010). In fact, environmental filtering could be identified with phylogeny as a proxy of functional differences among species at larger scales, but this pattern might have been caused by competitive exclusion leading to clustering after the elimination of the weakest competitors (Mayfield and Levine, 2010). It should be noted that this pattern was more obvious with NM1, which implies that the species pool selected passed a specific environmental filter, and thus the patterns detected could probably be attributed to the incidence of biotic factors (species competition for the same resources) rather than environmental heterogeneity (Stubbs and Wilson, 2004).
Functional Heterogeneity Across Spatial Scales
Overall, functional heterogeneity at the smallest scale (CV-CWMw) for the traits related to the leaf economic spectrum showed a variation similar to or even higher than that observed at the regional scale (CV-CWMr). These findings suggest the existence of (a) a primary abiotic filter at the regional scale determining low functional turnover throughout the distribution area and minor differences between plots (i.e., low CV-CWMr); (b) the existence of local environmental heterogeneity acting within each plot to yield functional heterogeneity (i.e., plots with relatively similar CV-CWMb values rather than CV-CWMr values); and (c) different and significant determinist processes at the smallest scale, which would also have been conditioning the species assembly (i.e., similar or higher CV-CWMw values to those between plots). As noted above, cold and summer drought are primary filters in Mediterranean high mountains that promote the existence of a similar and well-adapted species pool among sites (i.e., low composition and functional turnover along elevation; Helmus and Ives, 2012). This relatively limited pool of species (no more than 60 species along a 500-m elevation gradient in Sierra de Guadarrama National Park) may be subject to the effects of other sources of environmental heterogeneity (potential solar radiation, light competition, soil characteristics, and shrub encroachment) or simply to stochasticity. These sources could constrain the species assemblage in each plot and lead to functional heterogeneity between plots (i.e., CV-CWMr), which is also limited by the primary filters. Nevertheless, the ecological variables that lead to other sources of heterogeneity are relatively similar between the subplots in each plot (same elevation, potential solar radiation, or soil characteristics), thereby resulting in redundant species assemblages between subplots with similar functional heterogeneity (i.e., CV-CWMb). By contrast, at small spatial scales, the species in a plot are subject to additional assembly mechanisms based on micro-environmental gradients (Hutchings et al., 2003; Burton et al., 2011) or species interactions (Pescador et al., 2014), which may increase the functional heterogeneity at the cell scale. These additional assembly mechanisms lead to a scenario where the deterministic processes that operate at small scales are as important for generating functional variability as those observed at larger scales. In general, if the functional heterogeneity at the smallest scale is in the range of that at the regional scale, then any model that attempts to explore the species distribution and community composition based on the environmental conditions and species functional traits (see Shipley et al., 2006) should consider at least these sources of spatial variability.
Relevance of Ecological Factors on the Diversity Patterns
Linear averaged models showed that assembly processes were influenced by ecological factors that depended on the scale, functional traits, and null model considered (Mason et al., 2011; Spasojevic and Suding, 2012; de Bello et al., 2013a; Price et al., 2014; Reitalu et al., 2014). Species in plots located at the highest elevations exhibited a clustering pattern according to the multi-trait approach as well as for the SLA and seed mass traits. However, when the sampling granularity decreased to the cell level, phylogenetic overdispersion increased with the elevation but with no effects on functional responses. These results suggest that species at the plot level use a similar functional strategy to cope with the harsh conditions on the summits (Weiher and Keddy, 1995; Stubbs and Wilson, 2004; Schwilk and Ackerly, 2005; Chalmandrier et al., 2017). However, a phylogenetic overdispersion pattern is needed at the smallest scale to reduce competition with direct neighbors for the same local resources (Cavender-Bares et al., 2004, 2006; Pescador et al., 2014). Phylogenetic patterns have been investigated previously along elevation gradients with contrasting findings (Bryant et al., 2008; Culmsee and Leuschner, 2013; González-Caro et al., 2014; Li et al., 2014; Qian et al., 2014). In particular, similarly to our results, Bryant et al. (2008) found that plant communities tended to exhibit phylogenetic overdispersion at higher elevations, probably due to the prevalence and intensification of facilitation with elevation, whereas competition was more important at lower elevations. Furthermore, high levels of stress can lead to more open and discontinuous vegetation cover providing more gaps for colonization, and thus higher phylogenetic diversity (Reitalu et al., 2014). Based on the conceptual framework of functional and phylogenetic patterns (Webb et al., 2002; Cavender-Bares et al., 2004; Kraft and Ackerly, 2010), our findings imply that important traits for habitat specialization in harsh conditions probably exhibit convergent evolution (i.e., a low phylogenetic signal; Supplementary Table 7).
The absence of responses in terms of the functional and phylogenetic diversity to potential solar radiation at the plot and cell scales suggest that difference in the total annual radiation between north- and south-facing slopes was less important as a filter (Méndez-Toribio et al., 2017). However, multi-trait and seed clustering was detected at the subplot scale when the potential solar radiation values were higher. Thus, at this scale, co-occurring species must employ similar physiological and regenerative strategies to cope with water stress in plots with high potential solar radiation, especially in the Mediterranean-type climate. Irrespective of the spatial scale considered, the multiple effects of soil factors on functional patterns suggest high heterogeneity in the soil characteristics at small scales (Jackson and Caldwell, 1993; Hutchings et al., 2003; Gazol et al., 2013), especially given the strong effects of soil pH (Chytrý et al., 2003) and SOC in grasslands. Remarkably, we found clustering patterns for the traits related to the leaf economic spectrum and overdispersion patterns for Hmax and seed mass at higher pH values, and these patterns have not been described previously to the best of our knowledge. Furthermore, the prevalence of clustering as the species richness increases was found previously using functional (Carboni et al., 2014; Niu et al., 2014) and phylogenetic approaches (Reitalu et al., 2014). Thus, if the environment favors the co-occurrence of a great number of species, they will tend to be functionally and phylogenetically more redundant and similar (Cadotte et al., 2011), possibly because the regional species pool is unable to provide new candidates with different functional strategies to cope with harsh conditions (Yachi and Loreau, 2007; Pakeman, 2011). As a consequence, this predictor together with elevation had hardly any effect on the ecological clustering–overdispersion patterns when NM1 was used (i.e., the species pool was represented by the species present in the plot considered), probably because the species in null assemblages were from a species pool subject to the same elevation and species richness as the realized assemblage. Finally, the overdispersion patterns for the Hmax and FT traits were modulated by an increase in the functional heterogeneity at the smallest scale (CV-CWMw), and thus site-to-site functional variation should be considered as a relevant factor when assessing community assemblies.
Conclusion
Our multi-scale approach applied to a single plant community type with a low species turnover highlights the importance of evaluating functional and phylogenetic patterns at different spatial scales to understand the current community assembly. We showed that complex functional and phylogenetic patterns emerge depending on the spatial scale considered. Environmental filtering was found at larger spatial scales where species coexist under particular conditions, whereas functional overdispersion was present at small scales, thereby suggesting secondary niche differentiation after the effects of primary filters such as cold and summer drought. Phylogenetically based analysis indicated a pattern consistent with the functional analysis at larger scales, but this congruence was absent at the subplot and cell scales where the phylogenetic pattern remained clustered. Our results also demonstrated the importance of functional heterogeneity at the smallest scale in our plant community, thereby confirming the presence of primary environmental filters and suggesting the existence of additional assembly mechanisms based on micro-environmental gradients and species interactions at the smallest scale. Finally, our results show that functional and phylogenetic diversity patterns differ in terms of their responses to ecological factors across spatial scales, which highlights the need to develop adequate tools for predicting the response of vegetation to global warming (Reu et al., 2011) or the distributions of species based on the relationships between environmental conditions and species functional traits (Shipley et al., 2006). Given the shifts in the functional and phylogenetic responses along the elevation and among spatial scales, both sources of diversity should be considered by employing a multi-scale approach to help understand the future of alpine biodiversity under ongoing warming.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MW927222–MW927321. Data sets on vegetation co-occurence patterns, environmental variables, and functional traits for each plant species can be found in López-Angulo et al. (2019).
Author Contributions
DP, AE, and FV conceived and designed the study. DP and JL-A conducted the fieldwork. DP and FB performed the statistical analysis. DP wrote the manuscript. All authors contributed to the discussion and interpretation of data, revised the manuscript, and approved the submitted version.
Funding
This research was supported by the Spanish Ministry of Economy and Competitiveness (PHENOTYPES project, PGC2018-099115-B-100), Community of Madrid (REMEDINAL TE-CM project, S2018/EMT-4338), and Grant Agency of the Czech Republic (P505/12/1296).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Carlos Díaz Palomo, Felipe García García, and Jan Lepš for their technical and methodological assistance in this work. We would also like to thank Ana Millanes for help with phylogenetic reconstruction, Lars Markesteijn and Duncan E. Jackson for English language editing assistance, and staff at the Parque Nacional de la Sierra de Guadarrama for providing permission to work in the field area.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.622148/full#supplementary-material
References
Bernard-Verdier, M., Navas, M.-L., Vellend, M., Violle, C., Fayolle, A., and Garnier, E. (2012). Community assembly along a soil depth gradient: contrasting patterns of plant trait convergence and divergence in a Mediterranean rangeland. J. Ecol. 100, 1422–1433. doi: 10.1111/1365-2745.12003
Blomberg, S. P., Garland, T. Jr., and Ives, A. R. (2003). Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745. doi: 10.1554/0014-3820(2003)057[0717:tfpsic]2.0.co;2
Bryant, J. A., Lamanna, C., Morlon, H., Kerkhoff, A. J., Enquist, B. J., and Green, J. L. (2008). Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. U.S.A. 105, 11505–11511. doi: 10.1073/pnas.0801920105
Burnham, K. P., and Anderson, D. R. (2002). Model Selection and Multimodel Inference: a Practical Information-Theoretic Approach. New York, NY: Springer.
Burton, J. I., Mladenoff, D. J., Clayton, M. K., and Forrester, J. A. (2011). The roles of environmental filtering and colonization in the fine-scale spatial patterning of ground-layer plant communities in north temperate deciduous forests. J. Ecol. 99, 764–776. doi: 10.1111/j.1365-2745.2011.01807.x
Cadotte, M. W., Carscadden, K., and Mirotchnick, N. (2011). Beyond species: functional diversity and the maintenance of ecological processes and services. J. Appl. Ecol. 48, 1079–1087. doi: 10.1111/j.1365-2664.2011.02048.x
Carboni, M., de Bello, F., Janeček, Š, Doležal, J., Horník, J., Lepš, J., et al. (2014). Changes in trait divergence and convergence along a productivity gradient in wet meadows. Agric. Ecosyst. Environ. 182, 96–105. doi: 10.1016/j.agee.2013.12.014
Cavender-Bares, J., Ackerly, D., Baum, D., and Bazzaz, F. (2004). Phylogenetic overdispersion in Floridian oak communities. Am. Nat. 163, 823–843. doi: 10.1086/386375
Cavender-Bares, J., Keen, A., and Miles, B. (2006). Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology 87, 109–122.
Cavender-Bares, J., Kozak, K. H., Fine, P. V., and Kembel, S. W. (2009). The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715. doi: 10.1111/j.1461-0248.2009.01314.x
Chalmandrier, L., Münkemüller, T., Colace, M. P., Renaud, J., Aubert, S., Carlson, B. Z., et al. (2017). Spatial scale and intraspecific trait variability mediate assembly rules in alpine grasslands. J. Ecol. 105, 277–287. doi: 10.1111/1365-2745.12658
Chesson, P. (2000). Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. doi: 10.1146/annurev.ecolsys.31.1.343
Chytrý, M., Tichý, L., and Roleček, J. (2003). Local and regional patterns of species richness in Central European vegetation types along the pH/calcium gradient. Folia Geobot. 38, 429–442. doi: 10.1007/bf02803250
Cornelissen, J., Lavorel, S., Garnier, E., Diaz, S., Buchmann, N., Gurvich, D., et al. (2003). A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Austr. J. Bot. 51, 335–380. doi: 10.1071/bt02124
Culmsee, H., and Leuschner, C. (2013). Consistent patterns of elevational change in tree taxonomic and phylogenetic diversity across Malesian mountain forests. J. Biogeogr. 40, 1997–2010. doi: 10.1111/jbi.12138
de Bello, F., Carmona, C. P., Lepš, J., Szava-Kovats, R., and Pärtel, M. (2016). Functional diversity through the mean trait dissimilarity: resolving shortcomings with existing paradigms and algorithms. Oecologia 180, 933–940. doi: 10.1007/s00442-016-3546-0
de Bello, F., Lavorel, S., Lavergne, S., Albert, C. H., Boulangeat, I., Mazel, F., et al. (2013a). Hierarchical effects of environmental filters on the functional structure of plant communities: a case study in the French Alps. Ecography 36, 393–402. doi: 10.1111/j.1600-0587.2012.07438.x
de Bello, F., Vandewalle, M., Reitalu, T., Lepš, J., Prentice, H. C., Lavorel, S., et al. (2013b). Evidence for scale-and disturbance-dependent trait assembly patterns in dry semi-natural grasslands. J. Ecol. 101, 1237–1244. doi: 10.1111/1365-2745.12139
Díaz, S., Cabido, M., and Casanoves, F. (1998). Plant functional traits and environmental filters at a regional scale. J. Veg. Sci. 9, 113–122. doi: 10.2307/3237229
Drummond, A. J., Ho, S. Y., Phillips, M. J., and Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. doi: 10.1371/journal.pbio.0040088
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Emerson, B. C., and Gillespie, R. G. (2008). Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol. Evol. 23, 619–630. doi: 10.1016/j.tree.2008.07.005
Flynn, D. F., Mirotchnick, N., Jain, M., Palmer, M. I., and Naeem, S. (2011). Functional and phylogenetic diversity as predictors of biodiversity-ecosystem-function relationships. Ecology 92, 1573–1581. doi: 10.1890/10-1245.1
Freestone, A. L., and Inouye, B. D. (2006). Dispersal limitation and environmental heterogeneity shape scale-dependent diversity patterns in plant communities. Ecology 87, 2425–2432. doi: 10.1890/0012-9658(2006)87[2425:dlaehs]2.0.co;2
Freschet, G. T., Dias, A. T., Ackerly, D. D., Aerts, R., van Bodegom, P. M., Cornwell, W. K., et al. (2011). Global to community scale differences in the prevalence of convergent over divergent leaf trait distributions in plant assemblages. Glob. Ecol. Biogeogr. 20, 755–765. doi: 10.1111/j.1466-8238.2011.00651.x
Gandullo, J. (1974). Ensayo de evaluación cuantitativa de la insolación en función de la orientación y de la pendiente del terreno. An. INIA Ser. Recur. Nat. 1, 95–107.
Gazol, A., Tamme, R., Price, J. N., Hiiesalu, I., Laanisto, L., and Pärtel, M. (2013). A negative heterogeneity–diversity relationship found in experimental grassland communities. Oecologia 173, 545–555. doi: 10.1007/s00442-013-2623-x
Gerhold, P., Cahill, J. F., Winter, M., Bartish, I. V., and Prinzing, A. (2015). Phylogenetic patterns are not proxies of community assembly mechanisms (they are far better). Funct. Ecol. 29, 600–614. doi: 10.1111/1365-2435.12425
Giménez-Benavides, L., Escudero, A., and Iriondo, J. M. (2007). Local adaptation enhances seedling recruitment along an altitudinal gradient in a high mountain Mediterranean plant. Ann. Bot. 99, 723–734. doi: 10.1093/aob/mcm007
González-Caro, S., Umaña, M. N., Álvarez, E., Stevenson, P. R., and Swenson, N. G. (2014). Phylogenetic alpha and beta diversity in tropical tree assemblages along regional-scale environmental gradients in northwest South America. J. Plant Ecol. 7, 145–153. doi: 10.1093/jpe/rtt076
Gotelli, N. J. (2000). Null model analysis of species co-occurrence patterns. Ecology 81, 2606–2621. doi: 10.1890/0012-9658(2000)081[2606:nmaosc]2.0.co;2
Gotelli, N. J., and McCabe, D. J. (2002). Species co-occurrence: a meta-analysis of JM Diamond’s assembly rules model. Ecology 83, 2091–2096. doi: 10.1890/0012-9658(2002)083[2091:scoama]2.0.co;2
Götzenberger, L., de Bello, F., Bråthen, K. A., Davison, J., Dubuis, A., Guisan, A., et al. (2012). Ecological assembly rules in plant communities—approaches, patterns and prospects. Biol. Rev. 87, 111–127. doi: 10.1111/j.1469-185x.2011.00187.x
Hutchings, M. J., John, E. A., and Wijesinghe, D. K. (2003). Toward understanding the consequences of soil heterogeneity for plant populations and communities. Ecology 84, 2322–2334. doi: 10.1890/02-0290
Jackson, R., and Caldwell, M. (1993). The scale of nutrient heterogeneity around individual plants and its quantification with geostatistics. Ecology 74, 612–614. doi: 10.2307/1939320
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Körner, C. (2003). Alpine Plant Life: Functional Plant Ecology of High Mountain Ecosystems. Berlin: Springer-Verlag.
Kraft, N. J., and Ackerly, D. D. (2010). Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 80, 401–422. doi: 10.1890/09-1672.1
Kraft, N. J., Valencia, R., and Ackerly, D. D. (2008). Functional traits and niche-based tree community assembly in an Amazonian forest. Science 322, 580–582. doi: 10.1126/science.1160662
Laliberté, E., and Legendre, P. (2010). A distance-based framework for measuring functional diversity from multiple traits. Ecology 91, 299–305. doi: 10.1890/08-2244.1
Laliberté, E., and Shipley, B. (2011). FD: Measuring Functional Diversity from Multiple Traits, and Other Tools for Functional Ecology. R Package version 1.0.11.
Le Bagousse-Pinguet, Y., Liancourt, P., Götzenberger, L., de Bello, F., Altman, J., Brozova, V., et al. (2018). A multi-scale approach reveals random phylogenetic patterns at the edge of vascular plant life. Perspect. Plant Ecol. Evol. Syst. 30, 22–30. doi: 10.1016/j.ppees.2017.10.002
Leibold, M. A. (1998). Similarity and local co-existence of species in regional biotas. Evol. Ecol. 12, 95–110. doi: 10.1023/a:1006511124428
Levin, S. A. (1992). The problem of pattern and scale in ecology. Ecology 73, 1943–1967. doi: 10.2307/1941447
Li, X.-H., Zhu, X.-X., Niu, Y., and Sun, H. (2014). Phylogenetic clustering and overdispersion for alpine plants along elevational gradient in the Hengduan Mountains Region, southwest China. J. Syst. Evol. 52, 280–288. doi: 10.1111/jse.12027
López-Angulo, J., Pescador, D. S., Sánchez, A. M., Luzuriaga, A. L., Cavieres, L. A., and Escudero, A. (2019). Alpine vegetation dataset from three contrasting mountain ranges differing in climate and evolutionary history. Data Brief 27:104816. doi: 10.1016/j.dib.2019.104816
López-Angulo, J., de la Cruz, M., Pescador, D. S., Sánchez, A. M., and Escudero, A. (2021). A dimmer shade of pale: revealing the faint signature of local assembly processes on the structure of strongly filtered plant communities. Ecography 44, 87–97. doi: 10.1111/ecog.05405
López-Angulo, J., Swenson, N. G., Cavieres, L. A., and Escudero, A. (2018). Interactions between abiotic gradients determine functional and phylogenetic diversity patterns in Mediterranean-type climate mountains in the Andes. J. Veg. Sci. 29, 245–254. doi: 10.1111/jvs.12607
Losos, J. B. (1996). Phylogenetic perspectives on community ecology. Ecology 77, 1344–1354. doi: 10.2307/2265532
Luo, Y.-H., Liu, J., Tan, S.-L., Cadotte, M. W., Wang, Y.-H., Xu, K., et al. (2016). Trait-based community assembly along an elevational gradient in subalpine forests: quantifying the roles of environmental factors in inter-and intraspecific variability. PLoS One 11:e0155749. doi: 10.1371/journal.pone.0155749
MacArthur, R., and Levins, R. (1967). The limiting similarity, convergence, and divergence of coexisting species. Am. Nat. 101, 377–385. doi: 10.1086/282505
Maddison, D., and Maddison, W. (2003). MacClade, version 4.06 ed. Sunderland, MA: Sinauer Associates.
Mason, N., and Wilson, J. (2006). Mechanisms of species coexistence in a lawn community: mutual corroboration between two independent assembly rules. Commun. Ecol. 7, 109–116. doi: 10.1556/comec.7.2006.1.11
Mason, N. W., de Bello, F., Doležal, J., and Lepš, J. (2011). Niche overlap reveals the effects of competition, disturbance and contrasting assembly processes in experimental grassland communities. J. Ecol. 99, 788–796. doi: 10.1111/j.1365-2745.2011.01801.x
Mayfield, M. M., and Levine, J. M. (2010). Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 13, 1085–1093. doi: 10.1111/j.1461-0248.2010.01509.x
McCain, C. M., and Colwell, R. K. (2011). Assessing the threat to montane biodiversity from discordant shifts in temperature and precipitation in a changing climate. Ecol. Lett. 14, 1236–1245. doi: 10.1111/j.1461-0248.2011.01695.x
Méndez-Toribio, M., Ibarra-Manríquez, G., Navarrete-Segueda, A., and Paz, H. (2017). Topographic position, but not slope aspect, drives the dominance of functional strategies of tropical dry forest trees. Environ. Res. Lett. 12:085002. doi: 10.1088/1748-9326/aa717b
Messier, J., McGill, B. J., and Lechowicz, M. J. (2010). How do traits vary across ecological scales? A case for trait-based ecology. Ecol. Lett.. 13, 838–848.
Mouquet, N., Devictor, V., Meynard, C. N., Munoz, F., Bersier, L. F., Chave, J., et al. (2012). Ecophylogenetics: advances and perspectives. Biol. Rev. 87, 769–785. doi: 10.1111/j.1469-185x.2012.00224.x
Myers, J. A., Chase, J. M., Jiménez, I., Jørgensen, P. M., Araujo-Murakami, A., Paniagua-Zambrana, N., et al. (2013). Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecol. Lett. 16, 151–157. doi: 10.1111/ele.12021
Niu, K., Choler, P., de Bello, F., Mirotchnick, N., Du, G., and Sun, S. (2014). Fertilization decreases species diversity but increases functional diversity: a three-year experiment in a Tibetan alpine meadow. Agric. Ecosyst. Environ. 182, 106–112. doi: 10.1016/j.agee.2013.07.015
Olano, J. M., Almería, I., Eugenio, M., and Arx, G. (2013). Under pressure: how a Mediterranean high-mountain forb coordinates growth and hydraulic xylem anatomy in response to temperature and water constraints. Funct. Ecol. 27, 1295–1303. doi: 10.1111/1365-2435.12144
Pakeman, R. J. (2011). Functional diversity indices reveal the impacts of land use intensification on plant community assembly. J. Ecol. 99, 1143–1151. doi: 10.1111/j.1365-2745.2011.01853.x
Palmer, M. W. (1994). Variation in species richness: towards a unification of hypotheses. Folia Geobot. Phytotaxonomica 29, 511–530. doi: 10.1007/bf02883148
Pavoine, S., Vallet, J., Dufour, A. B., Gachet, S., and Daniel, H. (2009). On the challenge of treating various types of variables: application for improving the measurement of functional diversity. Oikos 118, 391–402. doi: 10.1111/j.1600-0706.2008.16668.x
Perronne, R., Mauchamp, L., Mouly, A., and Gillet, F. (2014). Contrasted taxonomic, phylogenetic and functional diversity patterns in semi-natural permanent grasslands along an altitudinal gradient. Plant Ecol. Evol. 147, 165–175. doi: 10.5091/plecevo.2014.885
Perronne, R., Munoz, F., Borgy, B., Reboud, X., and Gaba, S. (2017). How to design trait-based analyses of community assembly mechanisms: Insights and guidelines from a literature review. Perspect. Plant Ecol. Evol. Syst. 25, 29–44. doi: 10.1016/j.ppees.2017.01.004
Pescador, D. S., Chacón-Labella, J., Cruz, M., and Escudero, A. (2014). Maintaining distances with the engineer: patterns of coexistence in plant communities beyond the patch-bare dichotomy. New Phytol. 204, 140–148. doi: 10.1111/nph.12899
Pescador, D. S., de Bello, F., Valladares, F., and Escudero, A. (2015). Plant trait variation along an altitudinal gradient in Mediterranean high mountain grasslands: controlling the species turnover effect. PLoS One 10:e0118876. doi: 10.1371/journal.pone.0118876
Pescador, D. S., Sierra-Almeida, Á, Torres, P. J., and Escudero, A. (2016). Summer freezing resistance: a critical filter for plant community assemblies in Mediterranean high mountains. Front. Plant Sci. 7:194. doi: 10.3389/fpls.2016.00194
Pistón, N., Schöb, C., Armas, C., Prieto, I., and Pugnaire, F. I. (2016). Contribution of co-occurring shrub species to community richness and phylogenetic diversity along an environmental gradient. Perspect. Plant Ecol. Evol. Syst. 19, 30–39. doi: 10.1016/j.ppees.2016.02.002
Price, J. N., Gazol, A., Tamme, R., Hiiesalu, I., and Pärtel, M. (2014). The functional assembly of experimental grasslands in relation to fertility and resource heterogeneity. Funct. Ecol. 28, 509–519. doi: 10.1111/1365-2435.12186
Qian, H., Hao, Z., and Zhang, J. (2014). Phylogenetic structure and phylogenetic diversity of angiosperm assemblages in forests along an elevational gradient in Changbaishan, China. J. Plant Ecol. 7, 154–165. doi: 10.1093/jpe/rtt072
R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rambaut, A., Suchard, M., Xie, D., and Aj, D. (2014). Tracer v1.6. Available from http://beast.bio.ed.ac.uk/Tracer (accessed April 12, 2018).
Reitalu, T., Helm, A., Pärtel, M., Bengtsson, K., Gerhold, P., Rosén, E., et al. (2014). Determinants of fine-scale plant diversity in dry calcareous grasslands within the Baltic Sea region. Agric. Ecosyst. Environ. 182, 59–68. doi: 10.1016/j.agee.2012.11.005
Reu, B., Proulx, R., Bohn, K., Dyke, J. G., Kleidon, A., Pavlick, R., et al. (2011). The role of climate and plant functional trade-offs in shaping global biome and biodiversity patterns. Glob. Ecol. Biogeogr. 20, 570–581. doi: 10.1111/j.1466-8238.2010.00621.x
Schwilk, D. W., and Ackerly, D. D. (2005). Limiting similarity and functional diversity along environmental gradients. Ecol. Lett. 8, 272–281. doi: 10.1111/j.1461-0248.2004.00720.x
Seabloom, E. W., BJøRNSTAD, O. N., Bolker, B. M., and Reichman, O. J. (2005). Spatial signature of environmental heterogeneity, dispersal, and competition in successional grasslands. Ecol. Monogr. 75, 199–214. doi: 10.1890/03-0841
Shipley, B., Vile, D., and Garnier, É (2006). From plant traits to plant communities: a statistical mechanistic approach to biodiversity. Science 314, 812–814. doi: 10.1126/science.1131344
Siefert, A., Ravenscroft, C., Weiser, M. D., and Swenson, N. G. (2013). Functional beta-diversity patterns reveal deterministic community assembly processes in Eastern North American trees. Glob. Ecol. Biogeogr. 22, 682–691. doi: 10.1111/geb.12030
Spasojevic, M. J., and Suding, K. N. (2012). Inferring community assembly mechanisms from functional diversity patterns: the importance of multiple assembly processes. J. Ecol. 100, 652–661. doi: 10.1111/j.1365-2745.2011.01945.x
Stubbs, W. J., and Wilson, J. B. (2004). Evidence for limiting similarity in a sand dune community. J. Ecol. 92, 557–567. doi: 10.1111/j.0022-0477.2004.00898.x
Swenson, N. G., Enquist, B. J., Pither, J., Thompson, J., and Zimmerman, J. K. (2006). The problem and promise of scale dependency in community phylogenetics. Ecology 87, 2418–2424. doi: 10.1890/0012-9658(2006)87[2418:tpapos]2.0.co;2
Violle, C., Navas, M. L., Vile, D., Kazakou, E., Fortunel, C., Hummel, I., et al. (2007). Let the concept of trait be functional! Oikos 116, 882–892. doi: 10.1111/j.0030-1299.2007.15559.x
Webb, C. O. (2000). Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 156, 145–155. doi: 10.2307/3079215
Webb, C. O., Ackerly, D. D., McPeek, M. A., and Donoghue, M. J. (2002). Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448
Weiher, E., and Keddy, P. A. (1995). The assembly of experimental wetland plant communities. Oikos 73, 323–335. doi: 10.2307/3545956
Xu, J., Chen, Y., Zhang, L., Chai, Y., Wang, M., Guo, Y., et al. (2017). Using phylogeny and functional traits for assessing community assembly along environmental gradients: a deterministic process driven by elevation. Ecol. Evol. 7, 5056–5069. doi: 10.1002/ece3.3068
Yachi, S., and Loreau, M. (2007). Does complementary resource use enhance ecosystem functioning? A model of light competition in plant communities. Ecol. Lett. 10, 54–62. doi: 10.1111/j.1461-0248.2006.00994.x
Keywords: clustering, community assembly, elevation gradient, functional traits, overdispersion, phylogenetic distance, species diversity
Citation: Pescador DS, de Bello F, López-Angulo J, Valladares F and Escudero A (2021) Spatial Scale Dependence of Ecological Factors That Regulate Functional and Phylogenetic Assembly in a Mediterranean High Mountain Grassland. Front. Ecol. Evol. 9:622148. doi: 10.3389/fevo.2021.622148
Received: 30 October 2020; Accepted: 30 June 2021;
Published: 28 July 2021.
Edited by:
Charles K. Lee, University of Waikato, New ZealandReviewed by:
Zehao Shen, Peking University, ChinaZuzana Münzbergová, Academy of Sciences of the Czech Republic (ASCR), Czechia
Copyright © 2021 Pescador, de Bello, López-Angulo, Valladares and Escudero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David S. Pescador, david.sanchez@urjc.es
 David S. Pescador
David S. Pescador Francesco de Bello3
Francesco de Bello3  Jesús López-Angulo
Jesús López-Angulo Adrián Escudero
Adrián Escudero