Abstract
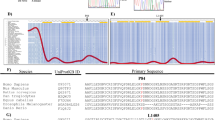
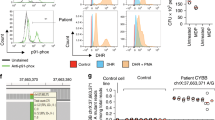
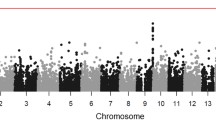
Genetic defects of innate immunity impairing intestinal bacterial sensing are linked to the development of Inflammatory Bowel Disease (IBD). Although much evidence supports a role of the intestinal virome in gut homeostasis, most studies focus on intestinal viral composition rather than on host intestinal viral sensitivity. To demonstrate the association between the development of Very Early Onset IBD (VEOIBD) and variants in the IFIH1 gene which encodes MDA5, a key cytosolic sensor for viral nucleic acids. Whole exome sequencing (WES) was performed in two independent cohorts of children with VEOIBD enrolled in Italy (n = 18) and USA (n = 24). Luciferase reporter assays were employed to assess MDA5 activity. An enrichment analysis was performed on IFIH1 comparing 42 VEOIBD probands with 1527 unrelated individuals without gastrointestinal or immunological issues. We identified rare, likely loss-of-function (LoF), IFIH1 variants in eight patients with VEOIBD from a combined cohort of 42 children. One subject, carrying a homozygous truncating variant resulting in complete LoF, experienced neonatal-onset, pan-gastrointestinal, IBD-like enteropathy plus multiple infectious episodes. The remaining seven subjects, affected by VEOIBD without immunodeficiency, were carriers of one LoF variant in IFIH1. Among these, two patients also carried a second hypomorphic variant, with partial function apparent when MDA5 was weakly stimulated. Furthermore, IFIH1 variants were significantly enriched in children with VEOIBD as compared to controls (p = 0.007). Complete and partial MDA5 deficiency is associated with VEOIBD with variable penetrance and expressivity, suggesting a role for impaired intestinal viral sensing in IBD pathogenesis.



Similar content being viewed by others
Abbreviations
- CD:
-
Crohn’s disease
- IBD:
-
Inflammatory bowel disease
- IFN:
-
Interferon
- LoF:
-
Loss-of-function
- SNVs:
-
Single nucleotide variants
- UC:
-
Ulcerative colitis
- IBDU:
-
Undetermined inflammatory bowel disease
- VEOIBD:
-
Very early-onset inflammatory bowel disease
- WES:
-
Whole exome sequencing
References
Asadpour-Behzadi A, Kariminik A (2018) RIG-1 and MDA5 are the important intracellular sensors against bacteria in septicemia suffering patients. J Appl Biomed 16:358–361
Asgari S et al. Loss-of-function mutations in IFIH1 predispose to severe viral respiratory infections in children, The Biology of Genomes Meeting. In: 2016.
Asgari S, Schlapbach LJ, Anchisi S et al (2017) Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc Natl Acad Sci U S A 114:8342–8347
Bainbridge MN, Wang M, Wu Y et al (2011) Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol 12:R68
Brisse M, Ly H (2019) Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front Immunol 10:1–27
Broggi A, Tan Y, Granucci F et al (2017) IFN-λ suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat Immunol 18:1084–1093
Broquet AH, Hirata Y, McAllister CS et al (2010) RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol 186:1618–1626
Ciancanelli MJ, Huang SXL, Luthra P et al (2015) Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 348(6233):448–453
Coban-Akdemir Z, White JJ, Song X et al (2018) Identifying genes whose mutant transcripts cause dominant disease traits by potential gain-of-function alleles. Am J Hum Genet 103:171–187
Coch C, Stümpel JP, Lilien-Waldau V et al (2017) RIG-I activation protects and rescues from lethal influenza virus infection and bacterial superinfection. Mol Ther 25:2093–2103
Corridoni D, Chapman T, Ambrose T et al (2018) Emerging mechanisms of innate immunity and their translational potential in inflammatory bowel disease. Front Med 5:1–22
Dai W, Liu Z-X, Li X-H et al (2007) Rig-I −/− mice develop colitis associated with downregulation of Gαi2. Cell Res 17:858–868
Depristo MA, Banks E, Poplin R et al (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–501
Derkach A, Lawless JF, Sun L (2013) Robust and powerful tests for rare variants using fisher’s method to combine evidence of association from two or more complementary tests. Genet Epidemiol 37:110–121
Dias Junior AG, Sampaio NG, Rehwinkel J (2019) A balancing act: MDA5 in antiviral immunity and autoinflammation. Trends Microbiol 27:75–85
Ellinghaus D, Jostins L, Spain SL et al (2016) Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 48:510–518
Enevold C, Oturai AB, Sørensen PS et al (2009) Multiple sclerosis and polymorphisms of innate pattern recognition receptors TLR1-10, NOD1-2, DDX58, and IFIH1. J Neuroimmunol 212:125–131
Fischer JC, Bscheider M, Eisenkolb G et al (2017) RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci Transl Med 9(386):eaag2513
Funke B, Lasitschka F, Roth W et al (2011) Selective downregulation of retinoic acid-inducible gene I within the intestinal epithelial compartment in Crohn’s disease. Inflamm Bowel Dis 17:1943–1954
Gorman JA, Hundhausen C, Errett JS et al (2017) The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat Immunol 18:744–752
Goubau D, Deddouche S (2013) Reis e sousa C. cytosolic sensing of viruses. Immunity 38:855–869
He L, Chen Y, Wu Y et al (2017) Nucleic acid sensing pattern recognition receptors in the development of colorectal cancer and colitis. Cell Mol Life Sci 74:2395–2411
Huang H, Fang M, Jostins L et al (2017) Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547:173–178
Hugot J, Chamaillard M, Zouali H (2001) Association of NOD2 leucine rich repeat variants with susceptibility to Crohn’s disease. Nature 411:599–603
Israel L, Wang Y, Bulek K et al (2017) Human adaptive immunity rescues an inborn error of innate immunity. Cell 168:789-800.e10
Itan Y, Mazel M, Mazel B et al (2014) HGCS: an online tool for prioritizing disease-causing gene variants by biological distance. BMC Genomics 15(1):256
Itan Y, Shang L, Boisson B et al (2015) The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci U S A 112:13615–13620
Itan Y, Shang L, Boisson B et al (2016) The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods 13:109–110
Kasumba DM, Grandvaux N (2019) Therapeutic targeting of RIG-I and MDA5 might not lead to the same rome. Trends Pharmacol Sci 40:116–127
Kato K, Ahmad S, Zhu Z et al (2020) Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. Mol Cell 117:1187
Kawaguchi S, Ishiguro Y, Imaizumi T et al (2009) Retinoic acid-inducible gene-I is constitutively expressed and involved in IFN-gamma-stimulated CXCL9-11 production in intestinal epithelial cells. Immunol Lett 123:9–13
Kelsen JR, Baldassano RN, Artis D et al (2015) Maintaining intestinal health: the genetics and immunology of very early onset inflammatory bowel disease. Cmgh 1:462–476
Kerur B, Benchimol EI, Fiedler K et al (2020) natural history of very early onset inflammatory bowel disease in north america: a retrospective cohort study. Inflamm Bowel Dis 27(3):295–302
Lamborn IT, Jing H, Zhang Y et al (2017) Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J Exp Med 214:1949–1972
Lee S, Baldridge MT (2017) Interferon-lambda: a potent regulator of intestinal viral infections. Front Immunol 8:749
Lee HC, Chathuranga K, Lee JS (2019) Intracellular sensing of viral genomes and viral evasion. Exp Mol Med 51(12):1–13
Levine A, Griffiths A, Markowitz J et al (2011) Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis 17:1314–1321
Levine A, Koletzko S, Turner D et al (2013) The ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J Pediatr Gastroenterol Nutr 58:1
Li H, Handsaker B, Wysoker A et al (2009) The sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079
Li X-D, Chiu Y-H, Ismail AS et al (2011) Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. Proc Natl Acad Sci 108:17390–17395
Li Q, Lee CH, Peters LA et al (2016) Variants in TRIM22 that affect NOD2 signaling are associated with very-early-onset inflammatory bowel disease. Gastroenterology 150:1196–1207
Lu Y, Li X, Liu S et al (2018) Toll-like receptors and inflammatory bowel disease. Front Immunol 9:1–9
Lupski JR, Gonzaga-Jauregui C, Yang Y et al (2013) Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med 5:57
MacDuff DA, Baldridge MT, Qaqish AM et al (2018) HOIL1 Is Essential for the Induction of Type I and III Interferons by MDA5 and Regulates Persistent Murine Norovirus Infection. J Virol 92(23):e01368-18
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Melchjorsen J, Rintahaka J, Søby S et al (2010) Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J Virol 84:11350–11358
Ori D, Murase M, Kawai T (2017) Cytosolic nucleic acid sensors and innate immune regulation. Int Rev Immunol 36:74–88
Parlato M, Charbit-Henrion F, Pan J et al (2018) Human ALPI deficiency causes inflammatory bowel disease and highlights a key mechanism of gut homeostasis. EMBO Mol Med 10:e8483
Pazmandi J, Kalinichenko A, Ardy RC et al (2019) Early-onset inflammatory bowel disease as a model disease to identify key regulators of immune homeostasis mechanisms. Immunol Rev 287:162–185
Pertea M, Lin X, Salzberg SL (2001) GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res 29:1185–1190
Picard C, von Bernuth H, Ghandil P et al (2010) Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (baltimore) 89:403–425
Reese MG, Eeckman FH, Kulp D et al (1997) Improved splice site detection in Genie. J Comput Biol 4:311–323
Rehwinkel J, Gack MU (2020) RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 20(9):537–551
Reid JG, Carroll A, Veeraraghavan N et al (2014) Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics 15:1–11
Ruel J, Ruane D, Mehandru S et al (2014) IBD across the age spectrum-Is it the same disease? Nat Rev Gastroenterol Hepatol 11:88–98
Rutsch F, MacDougall M, Lu C et al (2015) A specific IFIH1 gain-of-function mutation causes Singleton-Merten syndrome. Am J Hum Genet 96:275–282
Schrodinger L. The PyMOL Molecular Graphics System. 2010.
Schwerd T, Pandey S, Yang H-T et al (2017) Impaired antibacterial autophagy links granulomatous intestinal inflammation in Niemann-Pick disease type C1 and XIAP deficiency with NOD2 variants in Crohn’s disease. Gut 66:1060–1073
Schwerd T, Bryant RV, Pandey S et al (2018) NOX1 loss-of-function genetic variants in patients with inflammatory bowel disease. Mucosal Immunol 11:562–574
Smedley D, Robinson PN (2015) Phenotype-driven strategies for exome prioritization of human Mendelian disease genes. Genome Med 7:81
Smyth DJ, Cooper JD, Bailey R et al (2006) A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 38:617–619
Sobreira N, Schiettecatte F, Boehm C et al (2015) New tools for mendelian disease gene identification: phenoDB variant analysis module; and genematcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat 36:425–431
Solis M, Goubau D, Hiscott J (2007) RIG-I has guts: Identification of a role for RIG-I in colitis development. Cell Res 17:974–975
Uhlig HH (2013) Monogenic diseases associated with intestinal inflammation: implications for the understanding of inflammatory bowel disease. Gut 62:1795–1805
Uhlig HH, Schwerd T (2016) From genes to mechanisms. Inflamm Bowel Dis 22:202–212
Van EL, De SL, Pombal D et al (2015) Brief report: IFIH1 mutation causes systemic lupus erythematosus with selective IgA deficiency. Arthritis Rheumatol 67:1592–1597
Venselaar H, te Beek TAH, Kuipers RKP et al (2010) Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics 11(1):548
Webb K, Desguerre I, Ariaudo G et al (2014) Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet 46:503–509
Wu B, Peisley A, Richards C et al (2013) Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152:276–289
Yeo G, Burge CB (2004) Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11:377–394
Zaki M, Thoenes M, Kawalia A et al (2017) Recurrent and prolonged infections in a child with a homozygous IFIH1 nonsense mutation. Front Genet 8:1–6
Zeissig Y, Petersen B-S, Milutinovic S et al (2015) XIAP variants in male Crohn’s disease. Gut 64:66–76
Zhao Y, Karijolich J (2019) Know thyself: RIG-I-like receptor sensing of DNA virus infection. J Virol 93:1–22
Zhu H, Xu WY, Hu Z et al (2017) RNA virus receptor Rig-I monitors gut microbiota and inhibits colitis-associated colorectal cancer. J Exp Clin Cancer Res 36:1–11
Acknowledgements
The authors acknowledge the assistance of Ms. Helen Matthews. We thank the patients and their families for participating in the research study.
Funding
The work was funded by a grant from Fondazione Città della Speranza ONLUS, the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and the Baylor-Hopkins Center for Mendelian Genomics (NHGRI UM1 HG006542).
Author information
Authors and Affiliations
Contributions
MC, AM: wrote the manuscript and prepared the figures, which all authors reviewed. ALG, EW, HCS and NLMS: performed a critical review of the findings and participated in the preparation of the manuscript. MC, AM, PG, LB, MG, ALG: provided clinical care to the patients, collected and reviewed clinical data. CM: reviewed the histological specimens and prepared the figures. DC, MDC, AL: performed the genetic and the bioinformatic analyses for Italian cohort. EW, RM, NLMS, JEP, SNJ, JSF: performed the genetic analysis for the American cohort and the enrichment analysis. SB performed the connectome analysis. HJ, SB, WT, and YZ: planned and performed experiments to assess for variant function, with supervision by HCS, GP, AB, RK, HAK, MOE, JRL and DV: performed a critical review of the findings.
Corresponding authors
Ethics declarations
Conflict of interests
Baylor College of Medicine and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), formerly the Baylor Miraca Genetics Laboratories, which performs CMA and clinical ES. J.R.L. serves on the Scientific Advisory Board of the BG. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a coinventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. A.L. has a share in ownership of Research & Innovation (R&I Genetics) Srl. The other authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2 (MOV 21753 KB)
Supplementary file3 (MP4 848 KB)
Supplementary file4 (MP4 45445 KB)
Supplementary file5 (MP4 1079 KB)
Rights and permissions
About this article
Cite this article
Cananzi, M., Wohler, E., Marzollo, A. et al. IFIH1 loss-of-function variants contribute to very early-onset inflammatory bowel disease. Hum Genet 140, 1299–1312 (2021). https://doi.org/10.1007/s00439-021-02300-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02300-4