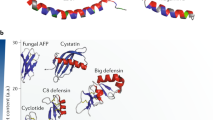
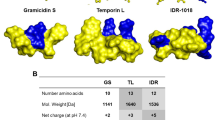
Abstract
Host defence peptides (HDPs) are integral components of innate immunity across all living organisms. These peptides can exert direct antibacterial effects, targeting planktonic cells (referred to as antimicrobial peptides), and exhibit antibiofilm (referred to as antibiofilm peptides), antiviral, antifungal and host-directed immunomodulatory activities. In this Review, we discuss how the complex functional attributes of HDPs provide many opportunities for the development of antimicrobial therapeutics, focusing particularly on their emerging antibiofilm properties. The mechanisms of action of antibiofilm peptides are compared and contrasted with those of antimicrobial peptides. Furthermore, obstacles for the practical translation of candidate peptides into therapeutics and the potential solutions are discussed. Critically, HDPs have the value-added assets of complex functional attributes, particularly antibiofilm and anti-inflammatory activities and their synergy with conventional antibiotics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fjell, C. D., Hiss, J. A., Hancock, R. E. W. & Schneider, G. Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 11, 37–51 (2012).
Nguyen, L. T., Haney, E. F. & Vogel, H. J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 29, 464–472 (2011).
Magana, M. et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 20, e216–e230 (2020). This Review identifies the benefits and opportunities of using antimicrobial peptides against multidrug-resistant pathogens, warranting the development of efficient developmental pipelines for their advancement.
Hancock, R. E. W., Haney, E. F. & Gill, E. E. The immunology of host defence peptides: beyond antimicrobial activity. Nat. Rev. Immunol. 16, 321–334 (2016). This Review highlights the immunomodulatory activities of host defence peptides beyond their antimicrobial activity.
Haney, E. F., Straus, S. K. & Hancock, R. E. W. Reassessing the host defense peptide landscape. Front. Chem. 7, 43 (2019).
Roope, L. S. J. et al. The challenge of antimicrobial resistance: what economics can contribute. Science 364, eaau4679 (2019).
Mookherjee, N., Anderson, M. A., Haagsman, H. P. & Davidson, D. J. Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discov. 19, 311–332 (2020). This Review focuses on the scope of functions of host defence peptides and emphasizes how diverse functionality is beneficial in clinical application.
Di, L. Strategic approaches to optimizing peptide ADME properties. AAPS J. 17, 134–143 (2014).
Hilchie, A. L., Wuerth, K. & Hancock, R. E. W. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat. Chem. Biol. 9, 761–768 (2013).
Patrzykat, A., Friedrich, C. L., Zhang, L., Mendoza, V. & Hancock, R. E. W. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 46, 605–614 (2002).
Kozlowska, J. et al. Combined systems approaches reveal highly plastic responses to antimicrobial peptide challenge in Escherichia coli. PLoS Pathog. 10, e1004104 (2014).
Majchrzykiewicz, J. A., Kuipers, O. P. & Bijlsma, J. J. E. Generic and specific adaptive responses of Streptococcus pneumoniae to challenge with three distinct antimicrobial peptides, bacitracin, LL-37, and nisin. Antimicrob. Agents Chemother. 54, 440–451 (2010).
Mensa, B., Howell, G. L., Scott, R. & DeGrado, W. F. Comparative mechanistic studies of brilacidin, daptomycin, and the antimicrobial peptide LL16. Antimicrob. Agents Chemother. 58, 5136–5145 (2014).
Joo, H. S., Fu, C. I. & Otto, M. Bacterial strategies of resistance to antimicrobial peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150292 (2016).
Bauer, M. E. & Shafer, W. M. On the in vivo significance of bacterial resistance to antimicrobial peptides. Biochim. Biophys. Acta 1848, 3101–3111 (2015).
Raetz, C. R. H., Reynolds, C. M., Trent, M. S. & Bishop, R. E. Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 (2007).
Shah, N. R., Hancock, R. E. W. & Fernandez, R. C. Bordetella pertussis lipid A glucosamine modification confers resistance to cationic antimicrobial peptides and increases resistance to outer membrane perturbation. Antimicrob. Agents Chemother. 58, 4931–4934 (2014).
Arroyo, L. A. et al. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 55, 3743–3751 (2011).
Moffatt, J. H. et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977 (2010).
Kristian, S. A. et al. d-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 187, 6719–6725 (2005).
Cox, E., Michalak, A., Pagentine, S., Seaton, P. & Pokorny, A. Lysylated phospholipids stabilize models of bacterial lipid bilayers and protect against antimicrobial peptides. Biochim. Biophys. Acta 1838, 2198–2204 (2014).
Mishra, N. N. & Bayer, A. S. Correlation of cell membrane lipid profiles with daptomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 1082–1085 (2013).
Schmidtchen, A., Frick, I. M., Andersson, E., Tapper, H. & Bjorck, L. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol 46, 157–168 (2002).
van der Plas, M. J. A. et al. Pseudomonas aeruginosa elastase cleaves a C-terminal peptide from human thrombin that inhibits host inflammatory responses. Nat. Commun. 7, 11567 (2016).
Jin, T. et al. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 172, 1169–1176 (2004).
Schmidtchen, A., Frick, I. M. & Björck, L. Dermatan sulphate is released by proteinases of common pathogenic bacteria and inactivates antibacterial α-defensin. Mol. Microbiol. 39, 708–713 (2001).
Manning, A. J. & Kuehn, M. J. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11, 258 (2011).
Band, V. I. & Weiss, D. S. Mechanisms of antimicrobial peptide resistance in Gram-negative bacteria. Antibiotics 4, 18–41 (2014).
Chakraborty, K. et al. Bacterial exotoxins downregulate cathelicidin (hCAP-18/LL-37) and human β-defensin 1 (HBD-1) expression in the intestinal epithelial cells. Cell. Microbiol. 10, 2520–2537 (2008).
Taggart, C. C. et al. Inactivation of human beta-defensins 2 and 3 by elastolytic cathepsins. J. Immunol. 171, 931–937 (2003).
Lazzaro, B. P., Zasloff, M. & Rolff, J. Antimicrobial peptides: application informed by evolution. Science 368, eaau5480 (2020).
Lázár, V. et al. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 3, 718–731 (2018). This study identifies multiple mutations that confer multidrug-resistance as well as collateral sensitivity to host defence peptides and shows that resistance development is more frequent to antibiotics than to peptides.
Spohn, R. et al. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 10, 4538 (2019).
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004).
Flemming, H. C. et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016).
Fernández, L., Breidenstein, E. B. M. & Hancock, R. E. W. Creeping baselines and adaptive resistance to antibiotics. Drug Resist. Updat. 14, 1–21 (2011).
Bryers, J. D. Medical Biofilms. Biotechnol. Bioeng. 100, 1–18 (2008).
Malone, M. et al. The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. J. Wound Care 26, 20–25 (2017).
Rumbaugh, K. P. & Sauer, K. Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586 (2020).
Kang, J., Dietz, M. J. & Li, B. Antimicrobial peptide LL-37 is bactericidal against Staphylococcus aureus biofilms. PLoS ONE 14, e0216676 (2019).
Overhage, J. et al. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 76, 4176–4182 (2008). This study was seminal to the field of biofilm-directed therapies as the first to identify the biofilm-specific effects of human cathelicidin LL-37.
Parducho, K. R. et al. The antimicrobial peptide human beta-defensin 2 inhibits biofilm production of Pseudomonas aeruginosa without compromising metabolic activity. Front. Immunol. 11, 805 (2020).
Zhu, C. et al. Human β-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation. J. Surg. Res. 183, 204–213 (2013).
Loutet, S. A. & Valvano, M. A. Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front. Microbiol. 2, 159 (2011).
de la Fuente-Núñez, C. et al. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 56, 2696–2704 (2012).
de la Fuente-Núñez, C., Reffuveille, F., Haney, E. F., Straus, S. K. & Hancock, R. E. W. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10, e1004152 (2014). This study was seminal to the field of biofilm-directed therapies as it identified a unique mechanism of action for a peptide with broad-spectrum activity.
Pletzer, D., Coleman, S. R. & Hancock, R. E. W. Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr. Opin. Microbiol. 33, 35–40 (2016). This Review summarizes peptides with biofilm-specific activities and describes, at least partially, their mechanism of action.
Hell, É., Giske, C. G., Nelson, A., Römling, U. & Marchini, G. Human cathelicidin peptide LL37 inhibits both attachment capability and biofilm formation of Staphylococcus epidermidis. Lett. Appl. Microbiol. 50, 211–215 (2010).
Pletzer, D. & Hancock, R. E. W. Antibiofilm peptides: potential as broad-spectrum agents. J. Bacteriol. 198, 2572–2578 (2016).
de la Fuente-Núñez, C. et al. D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 22, 196–205 (2015).
Mansour, S. C. et al. Bacterial abscess formation is controlled by the stringent stress response and can be targeted therapeutically. EBioMedicine 12, 219–226 (2016).
Pletzer, D., Wolfmeier, H., Bains, M. & Hancock, R. E. W. Synthetic peptides to target stringent response-controlled virulence in a Pseudomonas aeruginosa murine cutaneous infection model. Front. Microbiol. 8, 1867 (2017).
Libardo, M. D. J. et al. Nuclease activity gives an edge to host-defense peptide piscidin 3 over piscidin 1, rendering it more effective against persisters and biofilms. FEBS J. 284, 3662–3683 (2017). This study identifies the host defence peptide piscidin 3 as more active than piscidin 1 in the host milieu as nuclease exposure improves its biofilm-directed effects.
Okuda, K. et al. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob. Agents Chemother. 57, 5572–5579 (2013).
Pulido, D. et al. A novel RNase 3/ECP peptide for Pseudomonas aeruginosa biofilm eradication that combines antimicrobial, lipopolysaccharide binding, and cell-agglutinating activities. Antimicrob. Agents Chemother. 60, 6313–6325 (2016).
Liu, W. et al. Antimicrobial peptide Cec4 eradicates the bacteria of clinical carbapenem-resistant Acinetobacter baumannii biofilm. Front. Microbiol. 11, 1532 (2020).
Fabretti, F. et al. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 74, 4164–4171 (2006).
Lewenza, S. Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa. Front. Microbiol. 4, 21 (2013).
Chan, C., Burrows, L. L. & Deber, C. M. Helix induction in antimicrobial peptides by alginate in biofilms. J. Biol. Chem. 279, 38749–38754 (2004).
Vuong, C. et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6, 269–275 (2004).
Lin, L. et al. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant Gram-negative bacterial pathogens. EBioMedicine 2, 690–698 (2015).
Reffuveille, F., Fuente-Núñez, C., de la, Mansour, S. & Hancock, R. E. W. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 58, 5363–5371 (2014).
Pletzer, D., Mansour, S. C. & Hancock, R. E. W. Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog. 14, e1007084 (2018). This study demonstrates synergistic activities of peptides and antibiotics that are more effective at treating infections when used in combination.
Ruden, S. et al. Synergy pattern of short cationic antimicrobial peptides against multidrug-resistant Pseudomonas aeruginosa. Front. Microbiol. 10, 2740 (2019).
Koo, H. B. & Seo, J. Antimicrobial peptides under clinical investigation. Peptide Sci. 111, e24122 (2019).
Mahlapuu, M., Håkansson, J., Ringstad, L. & Björn, C. Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6, 194 (2016).
Hancock, R. E. W. & Sahl, H. G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557 (2006).
Meneguetti, B. T. et al. Antimicrobial peptides from fruits and their potential use as biotechnological tools — a review and outlook. Front. Microbiol. 7, 2136 (2017).
Pfalzgraff, A., Brandenburg, K. & Weindl, G. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front. Pharmacol. 9, 281 (2018).
Wu, B. et al. Human organoid biofilm model for assessing antibiofilm activity of novel agents. NPJ Biofilms Microbiomes 7, 1–13 (2021).
Lebeaux, D., Chauhan, A., Rendueles, O. & Beloin, C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2, 288–356 (2013).
Choi, K. Y. G., Wu, B. C., Lee, A. H. Y., Baquir, B. & Hancock, R. E. W. Utilizing organoid and air-liquid interface models as a screening method in the development of new host defense peptides. Front. Cell Infect. Microbiol. 10, 228 (2020).
Haney, E. F. et al. Identification of an IDR peptide formulation candidate that prevents peptide aggregation and retains immunomodulatory activity. Peptide Sci. 111, e24077 (2019).
Bos, J. D. & Meinardi, M. M. The 500 Dalton rule for the skin penetration of chemical compounds and drugs. Exp. Dermatol. 9, 165–169 (2000).
Bolouri, H. et al. Innate defense regulator peptide 1018 protects against perinatal brain injury. Ann Neurol. 75, 395–410 (2014).
de Breij, A. et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 10, eaan4044 (2018).
Huang, C. et al. Porcine beta-defensin 2 provides protection against bacterial infection by a direct bactericidal activity and alleviates inflammation via interference with the TLR4/NF-κB pathway. Front. Immunol. 10, 1673 (2019).
Wuerth, K., Lee, A. H. Y., Falsafi, R., Gill, E. E. & Hancock, R. E. W. Characterization of host responses during Pseudomonas aeruginosa acute lung infection in the lungs and blood and after treatment with the synthetic immunomodulatory peptide IDR-1002. Infect. Immun. 87, e00661-18 (2019).
Riquelme, S. A., Ahn, D. & Prince, A. Pseudomonas aeruginosa and Klebsiella pneumoniae adaptation to innate immune clearance mechanisms in the lung. J. Innate Immun. 10, 442–454 (2018).
Rivas-Santiago, B. et al. Ability of innate defence regulator peptides IDR-1002, IDR-HH2 and IDR-1018 to protect against Mycobacterium tuberculosis infections in animal models. PLoS ONE 8, e59119 (2013).
Chen, C., Deslouches, B., Montelaro, R. C. & Di, Y. P. Enhanced efficacy of the engineered antimicrobial peptide WLBU2 via direct airway delivery in a murine model of Pseudomonas aeruginosa pneumonia. Clin. Microbiol. Infect. 24, 547.e1–547.e8 (2018).
Di, Y. P. et al. Enhanced therapeutic index of an antimicrobial peptide in mice by increasing safety and activity against multidrug-resistant bacteria. Sci. Adv. 6, eaay6817 (2020).
Melvin, J. A. et al. Simultaneous antibiofilm and antiviral activities of an engineered antimicrobial peptide during virus-bacterium coinfection. mSphere 1, e00083-16 (2016).
Hoffmann, J. et al. Viral and bacterial co-infection in severe pneumonia triggers innate immune responses and specifically enhances IP-10: a translational study. Sci. Rep. 6, 38532 (2016).
Boisvert, A.-A., Cheng, M. P., Sheppard, D. C. & Nguyen, D. Microbial biofilms in pulmonary and critical care diseases. Ann. Am. Thorac. Soc. 13, 1615–1623 (2016).
Lebeaux, D., Ghigo, J. M. & Beloin, C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78, 510–543 (2014).
Haisma, E. M. et al. Antimicrobial peptide P60.4Ac-containing creams and gel for eradication of methicillin-resistant Staphylococcus aureus from cultured skin and airway epithelial surfaces. Antimicrob. Agents Chemother. 60, 4063–4072 (2016).
Peek, N. F. A. W. et al. Ototopical drops containing a novel antibacterial synthetic peptide: Safety and efficacy in adults with chronic suppurative otitis media. PLoS ONE 15, e0231573 (2020).
Riool, M., de Breij, A., Drijfhout, J. W., Nibbering, P. H. & Zaat, S. A. J. Antimicrobial peptides in biomedical device manufacturing. Front. Chem. 5, 63 (2017).
Yu, H. et al. Assessing the potential of four cathelicidins for the management of mouse candidiasis and Candida albicans biofilms. Biochimie 121, 268–277 (2016).
Freitas, C. G. et al. An immunomodulatory peptide confers protection in an experimental candidemia murine model. Antimicrob. Agents Chemother. 61, e02518-16 (2017).
Oshiro, K. G. N., Rodrigues, G., Monges, B. E. D., Cardoso, M. H. & Franco, O. L. Bioactive peptides against fungal biofilms. Front. Microbiol. 10, 2169 (2019).
Hao, H. et al. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 5, 288 (2014).
Stanford, K. et al. Antimicrobial resistance in members of the bacterial bovine respiratory disease complex isolated from lung tissue of cattle mortalities managed with or without the use of antimicrobials. Microorganisms 8, 288 (2020).
Kraemer, S. A., Ramachandran, A. & Perron, G. G. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms 7, 180 (2019).
Lim, S. J., Seo, C. K., Kim, T. H. & Myung, S. W. Occurrence and ecological hazard assessment of selected veterinary medicines in livestock wastewater treatment plants. J. Environ. Sci. Health B 48, 658–670 (2013).
Abdullahi, U. F., Igwenagu, E., Mu’azu, A., Aliyu, S. & Umar, M. I. Intrigues of biofilm: a perspective in veterinary medicine. Vet. World 9, 12–18 (2016).
Milivojevic, D. et al. Biofilm-forming ability and infection potential of Pseudomonas aeruginosa strains isolated from animals and humans. Pathog. Dis. 76, fty041 (2018).
Cabassi, C. S. et al. Activity of AMP2041 against human and animal multidrug resistant Pseudomonas aeruginosa clinical isolates. Ann. Clin. Microbiol. Antimicrob. 16, 17 (2017).
Greco, I. et al. In vitro ADME properties of two novel antimicrobial peptoid-based compounds as potential agents against canine pyoderma. Molecules 23, 630 (2018).
Greco, I. et al. Characterization, mechanism of action and optimization of activity of a novel peptide-peptoid hybrid against bacterial pathogens involved in canine skin infections. Sci. Rep. 9, 3679 (2019).
Danhorn, T. & Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61, 401–422 (2007).
Villa, F., Cappitelli, F., Cortesi, P. & Kunova, A. Fungal biofilms: targets for the development of novel strategies in plant disease management. Front. Microbiol. 8, 654 (2017).
Popp, J., Peto″, K. & Nagy, J. Pesticide productivity and food security: a review. Agron. Sustain. Dev. 33, 243–255 (2013).
Holaskova, E., Galuszka, P., Frebort, I. & Oz, M. T. Antimicrobial peptide production and plant-based expression systems for medical and agricultural biotechnology. Biotechnol. Adv. 33, 1005–1023 (2015). This Review summarizes plant-based expression systems of host defence peptides for preventing agricultural crop loss to pests and scaling up the production of peptides for clinical applications.
Sher Khan, R. et al. Plant defensins: types, mechanism of action and prospects of genetic engineering for enhanced disease resistance in plants. 3 Biotech 9, 192 (2019).
Theuretzbacher, U. & Piddock, L. J. V. Non-traditional antibacterial therapeutic options and challenges. Cell Host Microbe 26, 61–72 (2019).
Wu, M., Maier, E., Benz, R. & Hancock, R. E. W. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38, 7235–7242 (1999).
Pieta, P., Mirza, J. & Lipkowski, J. Direct visualization of the alamethicin pore formed in a planar phospholipid matrix. Proc. Natl Acad. Sci. USA 109, 21223–21227 (2012).
Fernandez, D. I. et al. The antimicrobial peptide aurein 1.2 disrupts model membranes via the carpet mechanism. Phys. Chem. Chem. Phys. 14, 15739–15751 (2012).
Leontiadou, H., Mark, A. E. & Marrink, S. J. Antimicrobial peptides in action. J. Am. Chem. Soc. 128, 12156–12161 (2006).
Park, C. B., Yi, K. S., Matsuzaki, K., Kim, M. S. & Kim, S. C. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc. Natl Acad. Sci. USA 97, 8245–8250 (2000).
Leveritt, J. M., Pino-Angeles, A. & Lazaridis, T. The structure of a melittin-stabilized pore. Biophys. J. 108, 2424–2426 (2015).
Gidalevitz, D. et al. Interaction of antimicrobial peptide protegrin with biomembranes. Proc. Natl Acad. Sci. USA 100, 6302–6307 (2003).
Schneider, T. et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 328, 1168–1172 (2010).
Andolina, G. et al. A peptidomimetic antibiotic interacts with the periplasmic domain of LptD from Pseudomonas aeruginosa. ACS Chem. Biol. 13, 666–675 (2018).
Cardon, S. et al. Peptidoglycan potentiates the membrane disrupting effect of the carboxyamidated form of DMS-DA6, a Gram-positive selective antimicrobial peptide isolated from Pachymedusa dacnicolor skin. PLoS ONE 13, e0205727 (2018).
Trimble, M. J., Mlynárcˇik, P., Kolárˇ, M. & Hancock, R. E. W. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 6, a025288 (2016).
Zhang, R. et al. Efficacy of antimicrobial peptide DP7, designed by machine-learning method, against methicillin-resistant Staphylococcus aureus. Front. Microbiol. 10, 1175 (2019).
Hilpert, K. et al. Short cationic antimicrobial peptides interact with ATP. Antimicrob. Agents Chemother. 54, 4480–4483 (2010).
Laughlin, T. F. & Ahmad, Z. Inhibition of Escherichia coli ATP synthase by amphibian antimicrobial peptides. Int. J. Biol. Macromol. 46, 367–374 (2010).
Di Somma, A. et al. The antimicrobial peptide Temporin L impairs E. coli cell division by interacting with FtsZ and the divisome complex. Biochim. Biophys. Acta 1864, 129606 (2020).
Ghosh, A. et al. Indolicidin targets duplex DNA: structural and mechanistic insight through a combination of spectroscopy and microscopy. ChemMedChem 9, 2052–2058 (2014).
Braffman, N. R. et al. Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin. Proc. Natl Acad. Sci. USA 116, 1273–1278 (2019).
Kragol, G. et al. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40, 3016–3026 (2001).
Seefeldt, A. C. et al. Structure of the mammalian antimicrobial peptide Bac7(1–16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 44, 2429–2438 (2016).
Wang, Y. et al. Antimicrobial peptide GH12 suppresses cariogenic virulence factors of Streptococcus mutans. J. Oral Microbiol. 10, 1442089 (2018).
Giacomucci, S., Cros, C. D. N., Perron, X., Mathieu-Denoncourt, A. & Duperthuy, M. Flagella-dependent inhibition of biofilm formation by sub-inhibitory concentration of polymyxin B in Vibrio cholerae. PLoS ONE 14, e0221431 (2019).
Brancatisano, F. L. et al. Inhibitory effect of the human liver-derived antimicrobial peptide hepcidin 20 on biofilms of polysaccharide intercellular adhesin (PIA)-positive and PIA-negative strains of Staphylococcus epidermidis. Biofouling 30, 435–446 (2014).
Hilpert, K., Volkmer-Engert, R., Walter, T. & Hancock, R. E. W. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 23, 1008–1012 (2005).
Tucker, A. T. et al. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell 172, 618–628.e13 (2018). This study describes a Surface Localized Antimicrobial Display platform for screening unlimited numbers of host defence peptides for a variety of functions, vastly increasing the number of known active peptide sequences.
Lee, E. Y., Lee, M. W., Fulan, B. M., Ferguson, A. L. & Wong, G. C. L. What can machine learning do for antimicrobial peptides, and what can antimicrobial peptides do for machine learning? Interface Focus 7, 20160153 (2017).
Etayash, H., Pletzer, D., Kumar, P., Straus, S. K. & Hancock, R. E. W. Cyclic derivative of host-defense peptide IDR-1018 improves proteolytic stability, suppresses inflammation, and enhances in vivo activity. J. Med. Chem. 63, 9228–9236 (2020).
Mourtada, R. et al. Design of stapled antimicrobial peptides that are stable, nontoxic and kill antibiotic-resistant bacteria in mice. Nat. Biotechnol. 37, 1186–1197 (2019). This study shows that stapled peptide derivatives are more effective and less toxic than naturally occurring peptides in a clinically relevant murine model of sepsis.
Luther, A. et al. Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 576, 452–458 (2019).
Carmona-Ribeiro, A. M. & Dias de Melo Carrasco, L. Novel formulations for antimicrobial peptides. Int. J. Mol. Sci. 15, 18040–18083 (2014).
Haney, E. F. et al. Computer-aided discovery of peptides that specifically attack bacterial biofilms. Sci. Rep. 8, 1871 (2018). This study describes a method for screening peptide libraries for antibiofilm activity and developing quantitative structure–activity relationship models for further optimization of screened peptides.
Xu, L. et al. Conversion of broad-spectrum antimicrobial peptides into species-specific antimicrobials capable of precisely targeting pathogenic bacteria. Sci. Rep. 10, 944 (2020).
Lemon, D. J. et al. Construction of a genetically modified T7Select phage system to express the antimicrobial peptide 1018. J. Microbiol. 57, 532–538 (2019).
Haney, E. F., Trimble, M. J., Cheng, J. T., Vallé, Q. & Hancock, R. E. W. Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules 8, 29 (2018).
Ceri, H. et al. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37, 1771–1776 (1999).
Locke, L. W. et al. Evaluation of peptide-based probes toward in vivo diagnostic imaging of bacterial biofilm-associated infections. ACS Infect. Dis. 6, 2086–2098 (2020).
Cieplik, F. et al. Microcosm biofilms cultured from different oral niches in periodontitis patients. J. Oral Microbiol. 11, 1551596 (2018).
Wang, Z., de la Fuente-Núñez, C., Shen, Y., Haapasalo, M. & Hancock, R. E. W. Treatment of oral multispecies biofilms by an anti-biofilm peptide. PLoS ONE 10, e0132512 (2015).
Zhang, T., Wang, Z., Hancock, R. E. W., de la Fuente-Núñez, C. & Haapasalo, M. Treatment of oral biofilms by a D-enantiomeric peptide. PLoS ONE 11, e0166997 (2016).
Huang, X. et al. Effect of long-term exposure to peptides on mono- and multispecies biofilms in dentinal tubules. J. Endod. 45, 1522–1528 (2019).
Jensen, L. K., Johansen, A. S. B. & Jensen, H. E. Porcine models of biofilm infections with focus on pathomorphology. Front. Microbiol. 8, 1961 (2017).
Khomtchouk, K. M. et al. A novel mouse model of chronic suppurative otitis media and its use in preclinical antibiotic evaluation. Sci. Adv. 6, eabc1828 (2020). This study describes a clinically relevant model of chronic suppurative otitis media (ear infection) and shows the importance of monitoring for recurrent infection when determining therapeutic efficacy.
Starr, C. G. et al. Synthetic molecular evolution of host cell-compatible, antimicrobial peptides effective against drug-resistant, biofilm-forming bacteria. Proc. Natl Acad. Sci. USA 117, 8437–8448 (2020).
Acknowledgements
Peptide research in the Hancock laboratory was recently supported by the Canadian Institutes of Health Research (CIHR) (Funding reference no. FDN-154287). M.A.A. is a Vanier and UBC Killam Doctoral Scholar and was supported by a Cystic Fibrosis Canada Studentship (#617081). R.E.W.H. holds a Canada Research Chair and is a UBC Killam Professor.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
E.F.H. and R.E.W.H. have filed patents related to the antibiofilm and immunomodulatory functions of synthetic HDPs. These patents have been assigned to their employer, the University of British Columbia, and licensed to ABT Innovations Inc., in which R.E.W.H. has an ownership position and E.F.H. owns shares. M.A.A. declares no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks G. Diamond, P. Stoodley and M. Zasloff for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Activity landscapes
-
The chemical spaces of peptides encompass all possible combinations of amino acids for a peptide of length N. The activity landscape can be conceptualized as a topographical map in which a particular function of a host defence peptide (for example, antimicrobial, antibiofilm or immune modulatory) is projected across all possible sequences within the peptide chemical space. In such an illustration, optimal peptide sequences would be represented by peaks, whereas less active peptides would localize within valleys.
- Minimal inhibitory concentration
-
(MIC). The lowest concentration of a compound that prevents visible growth of bacteria. Bacteria are considered susceptible to a compound if the MIC is below the clinical breakpoint for that compound.
- Twitching motility
-
A form of bacterial locomotion that depends on the type IV pilus in the model organism Pseudomonas aeruginosa and contributes to biofilm maturation as well as virulence.
- Stringent-stress response
-
A broadly conserved bacterial stress response that controls adaptation to nutrient deprivation and is activated by a number of different starvation and stress signals. The molecular hallmark of this response is synthesis of the small molecules guanosine tetraphosphate and its precursor, guanosine pentaphosphate.
- Abscess
-
A collection of pus within the tissues that can contain a high density of bacteria. Although not a biofilm per se, it shares several features with biofilms (local infection, can be chronic, high density, guanosine tetraphosphate dependence, adaptive antibiotic resistance or existence of matrix components).
- Biofilm matrix
-
The protective extracellular matrix of a biofilm consisting primarily of bacteria-produced molecules, including polysaccharides, proteins, lipids and extracellular DNA.
- Bacteriocin
-
Peptide toxins produced by bacteria that act upon closely related bacterial strains.
- Microcosm
-
A community, place or situation regarded as encapsulating, in miniature, the characteristic qualities or features of something large.
- Air–liquid interface (ALI) models
-
Cell culture technique in which mammalian cells are grown until they cover a support (for example, a membrane filter) and then the liquid on top of the cells is removed, enabling the growth of organ-like structures (for example, skin).
Rights and permissions
About this article
Cite this article
Hancock, R.E.W., Alford, M.A. & Haney, E.F. Antibiofilm activity of host defence peptides: complexity provides opportunities. Nat Rev Microbiol 19, 786–797 (2021). https://doi.org/10.1038/s41579-021-00585-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-021-00585-w
This article is cited by
-
Staphylococcus aureus and biofilms: transmission, threats, and promising strategies in animal husbandry
Journal of Animal Science and Biotechnology (2024)
-
Confronting the complexities of antimicrobial management for Staphylococcus aureus causing bovine mastitis: an innovative paradigm
Irish Veterinary Journal (2024)
-
Antibiotic Resistant Biofilms and the Quest for Novel Therapeutic Strategies
Indian Journal of Microbiology (2024)
-
Combining SNAPs with antibiotics shows enhanced synergistic efficacy against S. aureus and P. aeruginosa biofilms
npj Biofilms and Microbiomes (2023)
-
Improving antibacterial ability of Ti-Cu thin films with co-sputtering method
Scientific Reports (2023)